Project
Illumina Body Map 2
Navigation
Downloads
Results for VAX1_GSX2
Z-value: 0.77
Transcription factors associated with VAX1_GSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
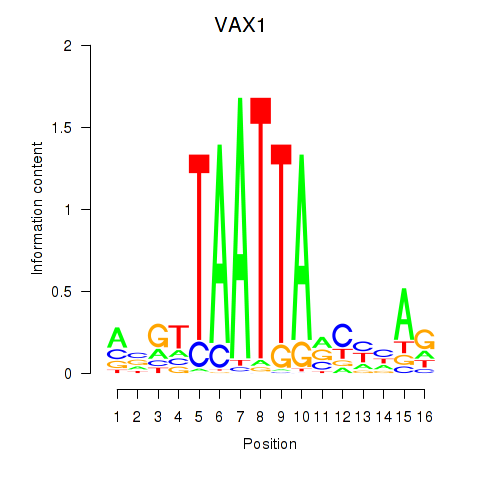
VAX1
|
ENSG00000148704.8 | ventral anterior homeobox 1 |
|
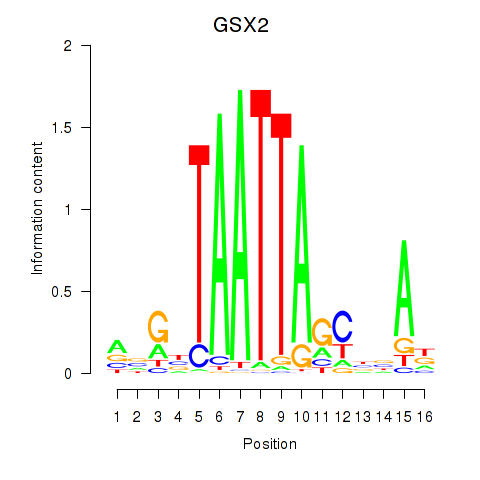
GSX2
|
ENSG00000180613.6 | GS homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VAX1 | hg19_v2_chr10_-_118897806_118897817 | 0.42 | 1.8e-02 | Click! |
| GSX2 | hg19_v2_chr4_+_54966198_54966340 | 0.41 | 1.9e-02 | Click! |
Activity profile of VAX1_GSX2 motif
Sorted Z-values of VAX1_GSX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_26593779 | 2.13 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr11_-_26593677 | 1.96 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr1_-_190446759 | 1.85 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr12_+_10163231 | 1.85 |
ENST00000396502.1
ENST00000338896.5 |
CLEC12B
|
C-type lectin domain family 12, member B |
| chr11_-_26593649 | 1.82 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr18_-_44181442 | 1.80 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr1_+_192778161 | 1.64 |
ENST00000235382.5
|
RGS2
|
regulator of G-protein signaling 2, 24kDa |
| chr13_+_32313658 | 1.64 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr1_-_185597619 | 1.61 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr17_+_61151306 | 1.55 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr11_+_65554493 | 1.51 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr3_-_164796269 | 1.44 |
ENST00000264382.3
|
SI
|
sucrase-isomaltase (alpha-glucosidase) |
| chr9_+_125132803 | 1.33 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_-_164534657 | 1.25 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr9_+_125133467 | 1.25 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr9_+_125133315 | 1.21 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr14_+_20187174 | 1.15 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr7_-_88425025 | 1.11 |
ENST00000297203.2
|
C7orf62
|
chromosome 7 open reading frame 62 |
| chr1_+_40713573 | 1.11 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr2_-_40680578 | 1.11 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr4_-_25865159 | 1.07 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr15_+_84115868 | 1.00 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr13_+_78315295 | 0.99 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_-_92952433 | 0.99 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr6_+_78400375 | 0.97 |
ENST00000602452.2
|
MEI4
|
meiosis-specific 4 homolog (S. cerevisiae) |
| chr3_+_111717511 | 0.96 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chrX_+_107288280 | 0.95 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_-_70326455 | 0.89 |
ENST00000374251.5
|
CXorf65
|
chromosome X open reading frame 65 |
| chr4_-_66536057 | 0.89 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr7_-_81399355 | 0.88 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr3_+_108541545 | 0.84 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr17_+_73539339 | 0.84 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr4_-_109541610 | 0.84 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr3_-_191000172 | 0.82 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr3_+_111718036 | 0.80 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr5_+_142286887 | 0.78 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr4_-_66536196 | 0.77 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr3_+_108541608 | 0.77 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr20_-_50419055 | 0.77 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr11_+_60163775 | 0.77 |
ENST00000300187.6
ENST00000395005.2 |
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr3_+_111717600 | 0.75 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr4_+_66536248 | 0.75 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr7_+_37723336 | 0.73 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr20_+_31805131 | 0.72 |
ENST00000375454.3
ENST00000375452.3 |
BPIFA3
|
BPI fold containing family A, member 3 |
| chr12_+_8666126 | 0.68 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr9_-_5339873 | 0.67 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr4_-_159956333 | 0.66 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr2_-_101925055 | 0.65 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr7_-_121944491 | 0.65 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr6_-_49931818 | 0.65 |
ENST00000322066.3
|
DEFB114
|
defensin, beta 114 |
| chr12_+_15699286 | 0.63 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr15_+_58430567 | 0.63 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr13_+_78315348 | 0.60 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr7_+_99202003 | 0.60 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr9_+_111624577 | 0.59 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr9_-_5304432 | 0.58 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr1_+_53925063 | 0.58 |
ENST00000371445.3
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr2_+_78143006 | 0.57 |
ENST00000443419.1
|
AC073628.1
|
AC073628.1 |
| chrX_+_107288239 | 0.56 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_+_45376922 | 0.55 |
ENST00000524410.1
ENST00000524488.1 ENST00000524565.1 |
RP11-430H10.1
|
RP11-430H10.1 |
| chr11_+_121447469 | 0.54 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr3_+_51851612 | 0.54 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr18_-_19997878 | 0.53 |
ENST00000391403.2
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr12_-_112123524 | 0.53 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr5_+_110559312 | 0.53 |
ENST00000508074.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr22_+_40297105 | 0.52 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr7_+_156433573 | 0.52 |
ENST00000311822.8
|
RNF32
|
ring finger protein 32 |
| chr6_-_33860521 | 0.52 |
ENST00000525746.1
ENST00000531046.1 |
LINC01016
|
long intergenic non-protein coding RNA 1016 |
| chr3_+_111718173 | 0.52 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr4_-_36245561 | 0.52 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr22_+_40297079 | 0.51 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr7_-_81399438 | 0.51 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chrX_+_107288197 | 0.49 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr19_-_52307357 | 0.49 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chrX_-_18690210 | 0.48 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr2_-_73520667 | 0.48 |
ENST00000545030.1
ENST00000436467.2 |
EGR4
|
early growth response 4 |
| chr15_+_63188009 | 0.48 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr15_+_75491203 | 0.48 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr3_+_44840679 | 0.47 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr5_-_96478466 | 0.47 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr18_-_67624160 | 0.47 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr2_-_166930131 | 0.46 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr20_-_50418972 | 0.46 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr3_+_111393501 | 0.45 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr6_+_155538093 | 0.44 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr7_-_81399329 | 0.44 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr11_-_102651343 | 0.44 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr7_-_81399411 | 0.43 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr10_-_92681033 | 0.43 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr14_+_39944025 | 0.43 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr21_-_15918618 | 0.42 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr16_+_58010339 | 0.41 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr14_+_72399833 | 0.41 |
ENST00000553530.1
ENST00000556437.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chrX_+_106765680 | 0.40 |
ENST00000276185.4
|
FRMPD3
|
FERM and PDZ domain containing 3 |
| chr3_+_140396881 | 0.40 |
ENST00000286349.3
|
TRIM42
|
tripartite motif containing 42 |
| chr4_-_112993808 | 0.40 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr9_+_95709733 | 0.40 |
ENST00000375482.3
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr7_+_37723450 | 0.39 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr10_+_91461413 | 0.39 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr6_-_87804815 | 0.39 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr6_+_127898312 | 0.39 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr7_-_35013217 | 0.38 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr10_-_28571015 | 0.38 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr2_+_3800169 | 0.38 |
ENST00000399143.3
|
DCDC2C
|
doublecortin domain containing 2C |
| chr5_+_148443049 | 0.38 |
ENST00000515304.1
ENST00000507318.1 |
CTC-529P8.1
|
CTC-529P8.1 |
| chr1_+_152974218 | 0.38 |
ENST00000331860.3
ENST00000443178.1 ENST00000295367.4 |
SPRR3
|
small proline-rich protein 3 |
| chr12_+_96196875 | 0.38 |
ENST00000553095.1
|
RP11-536G4.1
|
Uncharacterized protein |
| chr16_-_52061283 | 0.38 |
ENST00000566314.1
|
C16orf97
|
chromosome 16 open reading frame 97 |
| chr1_-_67266939 | 0.38 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr14_+_32798547 | 0.37 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr12_-_10607084 | 0.37 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr20_-_56647116 | 0.37 |
ENST00000441277.2
ENST00000452842.1 |
RP13-379L11.1
|
RP13-379L11.1 |
| chr11_+_59824060 | 0.36 |
ENST00000395032.2
ENST00000358152.2 |
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr12_-_10282742 | 0.36 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr14_+_22564294 | 0.36 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chr22_+_24990746 | 0.36 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr5_-_96478457 | 0.36 |
ENST00000512378.1
|
LIX1
|
Lix1 homolog (chicken) |
| chr13_-_41593425 | 0.36 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr11_-_36619771 | 0.36 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr3_+_62304648 | 0.35 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr21_-_22175341 | 0.35 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr7_-_92777606 | 0.35 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr14_+_105864847 | 0.35 |
ENST00000451127.2
|
TEX22
|
testis expressed 22 |
| chr5_-_147286065 | 0.35 |
ENST00000318315.4
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr3_+_62304712 | 0.35 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr15_-_75748115 | 0.35 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr19_+_17638059 | 0.35 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr6_+_13272904 | 0.35 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr14_-_57272366 | 0.34 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr11_-_27722021 | 0.34 |
ENST00000356660.4
ENST00000418212.1 ENST00000533246.1 |
BDNF
|
brain-derived neurotrophic factor |
| chr2_-_203735484 | 0.33 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr9_-_5830768 | 0.32 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr6_-_89927151 | 0.32 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr8_-_94179079 | 0.32 |
ENST00000521906.1
|
C8orf87
|
chromosome 8 open reading frame 87 |
| chr8_+_27238147 | 0.32 |
ENST00000412793.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr2_-_50201327 | 0.32 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chr15_-_64665911 | 0.32 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr1_-_68698197 | 0.32 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr11_-_133826852 | 0.31 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr14_+_22465771 | 0.31 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr6_+_46761118 | 0.31 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr17_-_73663245 | 0.31 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr17_-_29645836 | 0.31 |
ENST00000578584.1
|
CTD-2370N5.3
|
CTD-2370N5.3 |
| chr18_+_22040620 | 0.30 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr12_-_10282836 | 0.30 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr2_-_166060382 | 0.30 |
ENST00000409101.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chrY_+_24454970 | 0.30 |
ENST00000414629.2
ENST00000445779.2 |
RBMY1J
|
RNA binding motif protein, Y-linked, family 1, member J |
| chr11_-_13517565 | 0.29 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr1_-_197115818 | 0.29 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr19_-_7764960 | 0.29 |
ENST00000593418.1
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr17_-_57229155 | 0.29 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr9_+_90112767 | 0.28 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr18_-_67624412 | 0.28 |
ENST00000580335.1
|
CD226
|
CD226 molecule |
| chr3_-_112565703 | 0.28 |
ENST00000488794.1
|
CD200R1L
|
CD200 receptor 1-like |
| chr20_-_50418947 | 0.28 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr3_-_151034734 | 0.27 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chrX_-_77225135 | 0.27 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr4_-_110736505 | 0.27 |
ENST00000609440.1
|
RP11-602N24.3
|
RP11-602N24.3 |
| chr4_+_48492269 | 0.27 |
ENST00000327939.4
|
ZAR1
|
zygote arrest 1 |
| chr15_+_75491213 | 0.27 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr1_-_205325850 | 0.27 |
ENST00000537168.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr12_+_29376592 | 0.27 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr2_+_210518057 | 0.27 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr6_-_110501126 | 0.27 |
ENST00000368938.1
|
WASF1
|
WAS protein family, member 1 |
| chr1_+_117544366 | 0.27 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr17_-_38956205 | 0.26 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr8_-_86253888 | 0.26 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr2_+_210517895 | 0.26 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr10_-_135379132 | 0.26 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr7_+_29186192 | 0.26 |
ENST00000539406.1
|
CHN2
|
chimerin 2 |
| chr2_-_205099700 | 0.26 |
ENST00000419860.1
|
AC009498.1
|
AC009498.1 |
| chr18_-_67623906 | 0.26 |
ENST00000583955.1
|
CD226
|
CD226 molecule |
| chr12_+_66582919 | 0.26 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr12_+_29376673 | 0.25 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr5_+_174151536 | 0.25 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr2_+_169757750 | 0.25 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr20_+_42523336 | 0.25 |
ENST00000428529.1
|
RP5-1030M6.3
|
RP5-1030M6.3 |
| chr5_+_173930676 | 0.25 |
ENST00000504512.1
|
RP11-267A15.1
|
RP11-267A15.1 |
| chr1_-_68698222 | 0.25 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr12_-_10605929 | 0.24 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr9_+_5450503 | 0.24 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chr6_+_34204642 | 0.24 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr5_-_55412774 | 0.24 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr1_+_28199047 | 0.24 |
ENST00000373925.1
ENST00000328928.7 ENST00000373927.3 ENST00000427466.1 ENST00000442118.1 ENST00000373921.3 |
THEMIS2
|
thymocyte selection associated family member 2 |
| chr1_+_84630645 | 0.24 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_+_49121213 | 0.24 |
ENST00000548380.1
|
LINC00935
|
long intergenic non-protein coding RNA 935 |
| chr12_-_64062583 | 0.23 |
ENST00000542209.1
|
DPY19L2
|
dpy-19-like 2 (C. elegans) |
| chrX_-_13835147 | 0.23 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr8_-_86290333 | 0.23 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr11_-_795170 | 0.23 |
ENST00000481290.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr16_+_31885079 | 0.23 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr6_-_110501200 | 0.23 |
ENST00000392586.1
ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1
|
WAS protein family, member 1 |
| chr8_+_75262612 | 0.23 |
ENST00000220822.7
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr17_+_12569472 | 0.23 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr1_-_205325698 | 0.23 |
ENST00000460687.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr11_+_115498761 | 0.23 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr12_-_10282681 | 0.22 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chrX_+_37639264 | 0.22 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr1_+_84630574 | 0.22 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_+_33638033 | 0.22 |
ENST00000390389.3
|
TRBV23OR9-2
|
T cell receptor beta variable 23/OR9-2 (non-functional) |
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX1_GSX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.3 | 1.5 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 0.6 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 1.4 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 1.6 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.2 | 1.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 0.5 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 1.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.5 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.2 | 3.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 2.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 1.0 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.2 | 1.8 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.5 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.3 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 1.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.6 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.5 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.6 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.3 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.7 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.3 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.1 | 1.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 2.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.5 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.4 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.2 | GO:0090678 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.6 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 5.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.3 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.2 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 1.6 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.2 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.8 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.5 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 1.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.1 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 1.0 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.2 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0051549 | elastin metabolic process(GO:0051541) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.3 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 1.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 5.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 4.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 1.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 2.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 1.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 1.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 1.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.8 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.1 | 1.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.2 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.5 | GO:0005124 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.1 | 0.2 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.1 | 1.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 2.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 2.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 3.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 3.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 9.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 2.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |