Project
Illumina Body Map 2
Navigation
Downloads
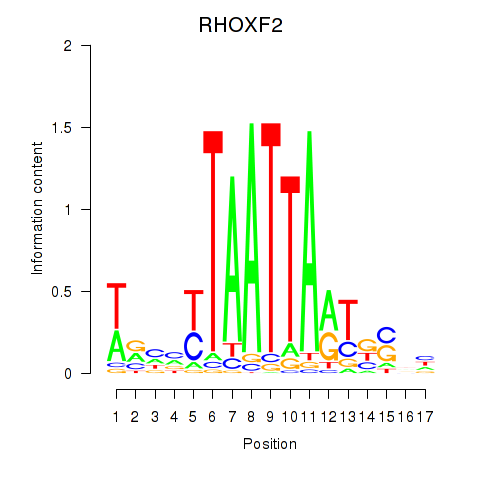
Results for VAX2_RHOXF2
Z-value: 0.75
Transcription factors associated with VAX2_RHOXF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VAX2
|
ENSG00000116035.2 | ventral anterior homeobox 2 |
|
RHOXF2
|
ENSG00000131721.4 | Rhox homeobox family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RHOXF2 | hg19_v2_chrX_+_119292467_119292467 | -0.49 | 4.0e-03 | Click! |
| VAX2 | hg19_v2_chr2_+_71127699_71127744 | 0.10 | 5.8e-01 | Click! |
Activity profile of VAX2_RHOXF2 motif
Sorted Z-values of VAX2_RHOXF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_89340242 | 3.03 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr2_+_90248739 | 2.83 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr9_+_125132803 | 2.23 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr9_+_125133315 | 2.08 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr8_+_92261516 | 1.76 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr6_+_26365443 | 1.64 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr6_+_26402517 | 1.60 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr7_+_50348268 | 1.55 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chrX_+_135730297 | 1.52 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr2_-_89292422 | 1.52 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr6_+_26365387 | 1.50 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr1_+_62439037 | 1.46 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr6_+_26402465 | 1.45 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr2_+_90043607 | 1.42 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr14_+_22564294 | 1.40 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chr15_+_96904487 | 1.37 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chrX_+_135730373 | 1.36 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr9_+_125133467 | 1.31 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_-_9760482 | 1.31 |
ENST00000229402.3
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1 |
| chr2_-_89597542 | 1.20 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr3_+_157154578 | 1.13 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr6_+_130339710 | 1.04 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr14_+_22670455 | 1.03 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr5_+_66300464 | 0.89 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr4_-_25865159 | 0.87 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr9_-_95166841 | 0.86 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr7_+_142495131 | 0.83 |
ENST00000390419.1
|
TRBJ2-7
|
T cell receptor beta joining 2-7 |
| chr7_+_115862858 | 0.75 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr4_+_169418195 | 0.74 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr10_-_92681033 | 0.74 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr10_-_17171817 | 0.74 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr4_-_74486217 | 0.73 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_+_40201954 | 0.69 |
ENST00000511121.1
|
RHOH
|
ras homolog family member H |
| chr12_+_131438496 | 0.68 |
ENST00000543826.1
|
GPR133
|
G protein-coupled receptor 133 |
| chr2_-_40680578 | 0.68 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr9_-_95166976 | 0.66 |
ENST00000447356.1
|
OGN
|
osteoglycin |
| chr4_+_169418255 | 0.66 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_-_74486347 | 0.66 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_66300446 | 0.65 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr16_+_12059050 | 0.63 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr4_-_74486109 | 0.63 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_-_92777606 | 0.62 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr17_-_40337470 | 0.62 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr14_+_22465771 | 0.60 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr12_+_131438443 | 0.60 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chrX_+_72783026 | 0.57 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr2_+_149402989 | 0.53 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr19_-_36001113 | 0.51 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr6_+_26440700 | 0.50 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr15_+_74218787 | 0.49 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr1_-_92952433 | 0.49 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_+_99717230 | 0.49 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr4_+_151503077 | 0.48 |
ENST00000317605.4
|
MAB21L2
|
mab-21-like 2 (C. elegans) |
| chr4_+_26324474 | 0.48 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_-_107729887 | 0.45 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr11_-_107729504 | 0.45 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr18_-_67624160 | 0.44 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr12_-_10022735 | 0.44 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr8_-_1922789 | 0.44 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr16_+_53133070 | 0.41 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_-_3469181 | 0.41 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr11_-_117747607 | 0.41 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr9_+_131062367 | 0.40 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr4_-_186696561 | 0.40 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr10_-_17171785 | 0.40 |
ENST00000377823.1
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr19_+_50016610 | 0.40 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_-_107729287 | 0.39 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr9_-_95166884 | 0.39 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr10_+_18549645 | 0.37 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr19_-_52307357 | 0.37 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr14_-_21994337 | 0.36 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chr2_-_214016314 | 0.35 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr11_-_117747434 | 0.34 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chrX_-_77225135 | 0.32 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr6_+_167536230 | 0.32 |
ENST00000341935.5
ENST00000349984.4 |
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr11_-_117748138 | 0.32 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr4_-_68829144 | 0.31 |
ENST00000508048.1
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr19_+_12175535 | 0.31 |
ENST00000550826.1
|
ZNF844
|
zinc finger protein 844 |
| chr2_+_169926047 | 0.28 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chrX_-_21676442 | 0.27 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr6_-_133035185 | 0.27 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr6_-_137494775 | 0.26 |
ENST00000349184.4
ENST00000296980.2 ENST00000339602.3 |
IL22RA2
|
interleukin 22 receptor, alpha 2 |
| chr9_-_5339873 | 0.26 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr7_-_99716914 | 0.26 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_+_142494920 | 0.25 |
ENST00000390418.1
|
TRBJ2-6
|
T cell receptor beta joining 2-6 |
| chr3_-_191000172 | 0.25 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr14_+_72399833 | 0.25 |
ENST00000553530.1
ENST00000556437.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr17_+_48823896 | 0.25 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr15_+_58430368 | 0.24 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr5_-_149829314 | 0.24 |
ENST00000407193.1
|
RPS14
|
ribosomal protein S14 |
| chr7_+_37723336 | 0.24 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr5_-_149829244 | 0.23 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chr16_-_28608364 | 0.23 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr18_-_67623906 | 0.23 |
ENST00000583955.1
|
CD226
|
CD226 molecule |
| chr10_+_99205894 | 0.22 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr15_-_37393406 | 0.22 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr4_-_186696425 | 0.22 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr13_+_36050881 | 0.21 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr14_+_101359265 | 0.21 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr4_-_138453559 | 0.21 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr6_-_33663474 | 0.21 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr12_+_26348246 | 0.20 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr1_+_225600404 | 0.20 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr17_-_41466555 | 0.18 |
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr15_-_98417780 | 0.18 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr11_-_62521614 | 0.17 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr14_-_20923195 | 0.17 |
ENST00000206542.4
|
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr1_+_160370344 | 0.17 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr10_+_90660832 | 0.17 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr19_+_12175504 | 0.16 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr15_+_58430567 | 0.16 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr4_-_186696515 | 0.16 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_121297650 | 0.16 |
ENST00000339397.4
|
SRFBP1
|
serum response factor binding protein 1 |
| chr3_+_152552685 | 0.16 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr9_+_12693336 | 0.15 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr17_-_39203519 | 0.15 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr1_+_46972668 | 0.14 |
ENST00000371956.4
ENST00000360032.3 |
DMBX1
|
diencephalon/mesencephalon homeobox 1 |
| chr4_-_69111401 | 0.14 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr4_+_159727272 | 0.14 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr10_+_99205959 | 0.14 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr12_-_50643664 | 0.13 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr10_-_50396407 | 0.13 |
ENST00000374153.2
ENST00000374151.3 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr3_+_158519654 | 0.13 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr7_+_148936732 | 0.13 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr14_+_56584414 | 0.12 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr20_-_33735070 | 0.12 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr7_-_44122063 | 0.12 |
ENST00000335195.6
ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM
|
polymerase (DNA directed), mu |
| chr1_+_54569968 | 0.12 |
ENST00000391366.1
|
AL161915.1
|
Uncharacterized protein |
| chr20_-_50722183 | 0.11 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr20_+_43990576 | 0.11 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr20_+_56136136 | 0.11 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr17_+_48823975 | 0.11 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr5_-_149829294 | 0.11 |
ENST00000401695.3
|
RPS14
|
ribosomal protein S14 |
| chr4_+_80584903 | 0.10 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr16_-_30122717 | 0.10 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr17_-_39191107 | 0.10 |
ENST00000344363.5
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr18_-_33709268 | 0.09 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr2_+_113763031 | 0.09 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr14_+_32547434 | 0.09 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr4_-_138453606 | 0.09 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr12_-_118796910 | 0.09 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr12_-_118797475 | 0.09 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr5_+_176811431 | 0.09 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr18_-_67624412 | 0.08 |
ENST00000580335.1
|
CD226
|
CD226 molecule |
| chr7_+_6655225 | 0.08 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr12_+_26348429 | 0.08 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr19_-_46088068 | 0.08 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr12_-_6233828 | 0.07 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr16_-_28608424 | 0.07 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr18_+_47087390 | 0.07 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr19_+_36632485 | 0.07 |
ENST00000586963.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr5_-_89770582 | 0.06 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr1_+_153747746 | 0.06 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr5_+_96840389 | 0.05 |
ENST00000504012.1
|
RP11-1E3.1
|
RP11-1E3.1 |
| chr14_+_67831576 | 0.05 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr15_+_49462434 | 0.05 |
ENST00000558145.1
ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2
|
galactokinase 2 |
| chr7_-_99716952 | 0.05 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr13_-_36050819 | 0.05 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr7_-_99717463 | 0.04 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr12_+_64798826 | 0.04 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr21_+_17443434 | 0.04 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr11_-_111794446 | 0.03 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr14_+_20923350 | 0.03 |
ENST00000555414.1
ENST00000216714.3 ENST00000553681.1 ENST00000557344.1 ENST00000398030.4 ENST00000557181.1 ENST00000555839.1 ENST00000553368.1 ENST00000556054.1 ENST00000557054.1 ENST00000557592.1 ENST00000557150.1 |
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr3_+_185431080 | 0.03 |
ENST00000296270.1
|
C3orf65
|
chromosome 3 open reading frame 65 |
| chr5_+_140557371 | 0.02 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr19_+_36632204 | 0.02 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr4_-_68829226 | 0.02 |
ENST00000396188.2
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr16_+_72088376 | 0.02 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr12_-_118796971 | 0.02 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr15_-_49912944 | 0.02 |
ENST00000559905.1
|
FAM227B
|
family with sequence similarity 227, member B |
| chr3_-_112127981 | 0.02 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr19_+_107471 | 0.02 |
ENST00000585993.1
|
OR4F17
|
olfactory receptor, family 4, subfamily F, member 17 |
| chr2_+_103089756 | 0.01 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr12_-_10955226 | 0.01 |
ENST00000240687.2
|
TAS2R7
|
taste receptor, type 2, member 7 |
| chr17_-_71258491 | 0.01 |
ENST00000397671.1
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr19_-_3557570 | 0.01 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr6_-_82957433 | 0.00 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr14_-_39639523 | 0.00 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr6_-_138866823 | 0.00 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX2_RHOXF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 6.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 0.5 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.2 | 1.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.8 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.5 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 2.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.6 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 1.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 1.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 0.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.5 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.2 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 1.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.2 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 7.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.8 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 5.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) glial limiting end-foot(GO:0097451) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.9 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.0 | 2.9 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 1.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.7 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 1.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.4 | GO:0005124 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.0 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 1.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 7.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0047035 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 5.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 3.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |