Project
Illumina Body Map 2
Navigation
Downloads
Results for VDR
Z-value: 1.13
Transcription factors associated with VDR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VDR
|
ENSG00000111424.6 | vitamin D receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VDR | hg19_v2_chr12_-_48276710_48276725 | -0.24 | 1.8e-01 | Click! |
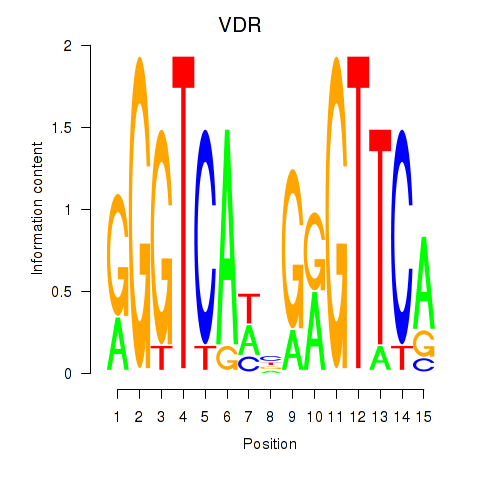
Activity profile of VDR motif
Sorted Z-values of VDR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_108905325 | 3.07 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr21_-_47575481 | 2.99 |
ENST00000291670.5
ENST00000397748.1 ENST00000359679.2 ENST00000355384.2 ENST00000397746.3 ENST00000397743.1 |
FTCD
|
formimidoyltransferase cyclodeaminase |
| chr17_-_64225508 | 2.87 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr9_+_139839711 | 2.83 |
ENST00000224181.3
|
C8G
|
complement component 8, gamma polypeptide |
| chr7_-_95025661 | 2.73 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr17_-_6616678 | 2.71 |
ENST00000381074.4
ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr11_-_116663127 | 2.64 |
ENST00000433069.1
ENST00000542499.1 |
APOA5
|
apolipoprotein A-V |
| chrX_+_70435044 | 2.45 |
ENST00000374029.1
ENST00000374022.3 ENST00000447581.1 |
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr17_-_7082861 | 2.42 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr9_-_136603456 | 2.38 |
ENST00000427237.2
ENST00000439388.1 |
SARDH
|
sarcosine dehydrogenase |
| chr14_+_94640633 | 2.20 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr14_+_95047744 | 2.11 |
ENST00000553511.1
ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr6_+_31916733 | 2.01 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr14_+_95048217 | 1.98 |
ENST00000557598.1
ENST00000556064.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr5_-_147211226 | 1.94 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr20_+_42193856 | 1.94 |
ENST00000412111.1
ENST00000423407.3 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr7_+_155090271 | 1.93 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr20_-_61992738 | 1.87 |
ENST00000370263.4
|
CHRNA4
|
cholinergic receptor, nicotinic, alpha 4 (neuronal) |
| chr7_+_100318423 | 1.86 |
ENST00000252723.2
|
EPO
|
erythropoietin |
| chr2_+_108905095 | 1.84 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr7_+_155089486 | 1.74 |
ENST00000340368.4
ENST00000344756.4 ENST00000425172.1 ENST00000342407.5 |
INSIG1
|
insulin induced gene 1 |
| chr3_+_190105909 | 1.68 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr9_+_139839686 | 1.63 |
ENST00000371634.2
|
C8G
|
complement component 8, gamma polypeptide |
| chr7_+_29234101 | 1.59 |
ENST00000435288.2
|
CHN2
|
chimerin 2 |
| chr5_-_147211190 | 1.55 |
ENST00000510027.2
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr16_+_20685815 | 1.52 |
ENST00000561584.1
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr12_-_96390108 | 1.52 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr7_+_29234028 | 1.50 |
ENST00000222792.6
|
CHN2
|
chimerin 2 |
| chr2_-_89160425 | 1.50 |
ENST00000390239.2
|
IGKJ4
|
immunoglobulin kappa joining 4 |
| chr19_-_3478477 | 1.49 |
ENST00000591708.1
|
C19orf77
|
chromosome 19 open reading frame 77 |
| chr4_+_158142750 | 1.46 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr6_-_25874440 | 1.40 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr2_-_89161064 | 1.39 |
ENST00000390241.2
|
IGKJ2
|
immunoglobulin kappa joining 2 |
| chr17_+_68100989 | 1.39 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_-_96390063 | 1.35 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr21_+_45905448 | 1.33 |
ENST00000449713.1
|
AP001065.15
|
AP001065.15 |
| chr19_-_16045619 | 1.27 |
ENST00000402119.4
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr19_-_16045665 | 1.24 |
ENST00000248041.8
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr12_-_96389702 | 1.17 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr7_+_97840739 | 1.16 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15 |
| chr4_+_85504075 | 1.15 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr3_-_27763803 | 1.13 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr22_+_35776354 | 1.04 |
ENST00000412893.1
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr16_-_68404908 | 0.99 |
ENST00000574662.1
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr8_+_9183618 | 0.99 |
ENST00000518619.1
|
RP11-115J16.1
|
RP11-115J16.1 |
| chr1_-_205325698 | 0.97 |
ENST00000460687.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr20_+_36974759 | 0.97 |
ENST00000217407.2
|
LBP
|
lipopolysaccharide binding protein |
| chr13_+_24844819 | 0.97 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr1_-_205325850 | 0.93 |
ENST00000537168.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr17_-_79805146 | 0.90 |
ENST00000415593.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr1_-_75198940 | 0.89 |
ENST00000417775.1
|
CRYZ
|
crystallin, zeta (quinone reductase) |
| chr1_-_205325994 | 0.88 |
ENST00000491471.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr3_-_27764190 | 0.88 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr13_+_24844857 | 0.88 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr1_-_205326161 | 0.87 |
ENST00000367156.3
ENST00000606887.1 ENST00000607173.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr1_-_205326022 | 0.86 |
ENST00000367155.3
|
KLHDC8A
|
kelch domain containing 8A |
| chr2_-_89160770 | 0.82 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr5_+_175487692 | 0.80 |
ENST00000510151.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr19_+_18077881 | 0.80 |
ENST00000609922.1
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr12_+_79439461 | 0.80 |
ENST00000552624.1
|
SYT1
|
synaptotagmin I |
| chr7_+_1272522 | 0.78 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr7_-_36764062 | 0.78 |
ENST00000435386.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr19_-_22715280 | 0.75 |
ENST00000593802.1
|
ZNF98
|
zinc finger protein 98 |
| chr1_-_67519782 | 0.73 |
ENST00000235345.5
|
SLC35D1
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
| chr11_+_60869980 | 0.73 |
ENST00000544014.1
|
CD5
|
CD5 molecule |
| chr7_-_96132835 | 0.72 |
ENST00000356686.1
|
C7orf76
|
chromosome 7 open reading frame 76 |
| chr14_+_21498360 | 0.71 |
ENST00000321760.6
ENST00000460647.2 ENST00000530140.2 ENST00000472458.1 |
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chr13_+_24844979 | 0.71 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr18_-_6414884 | 0.70 |
ENST00000317931.7
ENST00000284898.6 ENST00000400104.3 |
L3MBTL4
|
l(3)mbt-like 4 (Drosophila) |
| chr8_-_8046331 | 0.70 |
ENST00000594215.1
|
LRLE1
|
Liver-related low express protein 1; Uncharacterized protein |
| chr12_+_79439405 | 0.70 |
ENST00000552744.1
|
SYT1
|
synaptotagmin I |
| chr12_-_133050726 | 0.68 |
ENST00000595994.1
|
MUC8
|
mucin 8 |
| chr10_-_18940501 | 0.66 |
ENST00000377304.4
|
NSUN6
|
NOP2/Sun domain family, member 6 |
| chr4_-_36246060 | 0.65 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_-_36764004 | 0.65 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr5_-_7869108 | 0.64 |
ENST00000264669.5
ENST00000507572.1 ENST00000504695.1 |
FASTKD3
|
FAST kinase domains 3 |
| chr2_+_220363579 | 0.63 |
ENST00000313597.5
ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chrX_-_73061339 | 0.62 |
ENST00000602863.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chrX_+_153770421 | 0.62 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr7_-_36764142 | 0.61 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr18_-_48351743 | 0.61 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr3_+_46618727 | 0.61 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr17_+_53828333 | 0.60 |
ENST00000268896.5
|
PCTP
|
phosphatidylcholine transfer protein |
| chr7_+_65552756 | 0.60 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr5_-_39270725 | 0.60 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr11_-_111320706 | 0.59 |
ENST00000531398.1
|
POU2AF1
|
POU class 2 associating factor 1 |
| chr14_-_101351184 | 0.59 |
ENST00000534062.1
|
RTL1
|
retrotransposon-like 1 |
| chr11_-_7904464 | 0.57 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr11_-_61582579 | 0.57 |
ENST00000539419.1
ENST00000545245.1 ENST00000545405.1 ENST00000542506.1 |
FADS1
|
fatty acid desaturase 1 |
| chr16_-_31105870 | 0.56 |
ENST00000394971.3
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr4_-_36245561 | 0.56 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_63258261 | 0.55 |
ENST00000506598.1
|
HTR1A
|
5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled |
| chr5_-_177210399 | 0.55 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr7_+_129015484 | 0.54 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr1_+_198608292 | 0.54 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_11752379 | 0.53 |
ENST00000396123.1
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr20_+_48909240 | 0.53 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr12_-_25801478 | 0.53 |
ENST00000540106.1
ENST00000445693.1 ENST00000545543.1 ENST00000542224.1 |
IFLTD1
|
intermediate filament tail domain containing 1 |
| chr11_-_15643937 | 0.53 |
ENST00000533082.1
|
RP11-531H8.2
|
RP11-531H8.2 |
| chr8_-_73793975 | 0.53 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr11_+_113779704 | 0.53 |
ENST00000537778.1
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr9_+_131644781 | 0.52 |
ENST00000259324.5
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr16_-_31106048 | 0.51 |
ENST00000300851.6
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr1_-_99470368 | 0.50 |
ENST00000263177.4
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr10_-_134756030 | 0.50 |
ENST00000368586.5
ENST00000368582.2 |
TTC40
|
tetratricopeptide repeat domain 40 |
| chr19_-_11494975 | 0.50 |
ENST00000222139.6
ENST00000592375.2 |
EPOR
|
erythropoietin receptor |
| chr16_-_2260834 | 0.49 |
ENST00000562360.1
ENST00000566018.1 |
BRICD5
|
BRICHOS domain containing 5 |
| chr12_+_110011571 | 0.49 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr16_-_31106211 | 0.49 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1
VKORC1
|
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr14_+_101359265 | 0.49 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr6_+_131521120 | 0.49 |
ENST00000537868.1
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr4_-_39640700 | 0.49 |
ENST00000295958.5
|
SMIM14
|
small integral membrane protein 14 |
| chr17_+_53828381 | 0.48 |
ENST00000576183.1
|
PCTP
|
phosphatidylcholine transfer protein |
| chr12_-_63328817 | 0.48 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chrX_+_149737046 | 0.47 |
ENST00000370396.2
ENST00000542741.1 ENST00000543350.1 ENST00000424519.1 ENST00000413012.2 |
MTM1
|
myotubularin 1 |
| chr1_-_48227290 | 0.47 |
ENST00000594280.1
|
FLJ00388
|
FLJ00388 |
| chr1_+_200993071 | 0.46 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr5_-_179227540 | 0.46 |
ENST00000520875.1
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr2_+_38893208 | 0.46 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr10_-_43892668 | 0.46 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr18_-_6414797 | 0.45 |
ENST00000583809.1
|
L3MBTL4
|
l(3)mbt-like 4 (Drosophila) |
| chr4_-_164534657 | 0.45 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr5_+_177433406 | 0.45 |
ENST00000504530.1
|
FAM153C
|
family with sequence similarity 153, member C |
| chr14_+_22978168 | 0.45 |
ENST00000390505.1
|
TRAJ32
|
T cell receptor alpha joining 32 |
| chr10_-_43892279 | 0.45 |
ENST00000443950.2
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr2_+_230787201 | 0.45 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr11_-_14992712 | 0.45 |
ENST00000486207.1
|
CALCA
|
calcitonin-related polypeptide alpha |
| chr16_-_30023615 | 0.45 |
ENST00000564979.1
ENST00000563378.1 |
DOC2A
|
double C2-like domains, alpha |
| chr20_+_3801162 | 0.44 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr19_+_49122548 | 0.43 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr1_-_200379180 | 0.43 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr4_-_103266626 | 0.43 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr1_-_221915418 | 0.43 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chrX_+_36254051 | 0.43 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr12_+_52626898 | 0.43 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr4_-_23735183 | 0.43 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr9_-_139343294 | 0.42 |
ENST00000313084.5
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr11_-_104817919 | 0.42 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_+_230787213 | 0.42 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr11_+_60869867 | 0.42 |
ENST00000347785.3
|
CD5
|
CD5 molecule |
| chr4_-_103266219 | 0.42 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr19_+_49122785 | 0.41 |
ENST00000598088.1
|
SPHK2
|
sphingosine kinase 2 |
| chr2_+_38893047 | 0.40 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr1_+_153750622 | 0.40 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr7_+_2685164 | 0.40 |
ENST00000400376.2
|
TTYH3
|
tweety family member 3 |
| chr19_-_54804173 | 0.40 |
ENST00000391744.3
ENST00000251390.3 |
LILRA3
|
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
| chr8_+_119294456 | 0.40 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr22_-_31536480 | 0.39 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chr9_+_140125209 | 0.39 |
ENST00000538474.1
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr14_-_54908043 | 0.38 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr12_+_53895364 | 0.38 |
ENST00000552817.1
ENST00000394357.2 |
TARBP2
|
TAR (HIV-1) RNA binding protein 2 |
| chr14_+_22591276 | 0.37 |
ENST00000390455.3
|
TRAV26-1
|
T cell receptor alpha variable 26-1 |
| chr5_-_177209832 | 0.37 |
ENST00000393518.3
ENST00000505531.1 ENST00000503567.1 |
FAM153A
|
family with sequence similarity 153, member A |
| chr14_+_77244349 | 0.36 |
ENST00000554743.1
|
VASH1
|
vasohibin 1 |
| chr19_+_16830815 | 0.36 |
ENST00000549814.1
|
NWD1
|
NACHT and WD repeat domain containing 1 |
| chr2_-_32489922 | 0.36 |
ENST00000402280.1
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr1_-_200992827 | 0.36 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr5_+_177433973 | 0.36 |
ENST00000507848.1
|
FAM153C
|
family with sequence similarity 153, member C |
| chr12_-_56848426 | 0.35 |
ENST00000257979.4
|
MIP
|
major intrinsic protein of lens fiber |
| chr3_-_122283079 | 0.35 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr1_-_99470558 | 0.35 |
ENST00000370188.3
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr5_+_175488258 | 0.35 |
ENST00000512862.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr20_+_2276639 | 0.35 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr1_+_198608146 | 0.35 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr17_+_53828432 | 0.34 |
ENST00000573500.1
|
PCTP
|
phosphatidylcholine transfer protein |
| chr20_-_52790512 | 0.34 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr3_+_184032919 | 0.33 |
ENST00000427845.1
ENST00000342981.4 ENST00000319274.6 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr14_-_75330537 | 0.33 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr15_-_81282133 | 0.33 |
ENST00000261758.4
|
MESDC2
|
mesoderm development candidate 2 |
| chr14_+_22984601 | 0.33 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr14_+_100531615 | 0.32 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr2_-_73511559 | 0.32 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr4_+_146601356 | 0.32 |
ENST00000438731.1
ENST00000511965.1 |
C4orf51
|
chromosome 4 open reading frame 51 |
| chr16_+_69958887 | 0.32 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr22_-_29663954 | 0.32 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chr3_-_122283100 | 0.31 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr4_-_122302163 | 0.31 |
ENST00000394427.2
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr14_+_58797974 | 0.31 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr14_+_100531738 | 0.31 |
ENST00000555706.1
|
EVL
|
Enah/Vasp-like |
| chr17_-_36348610 | 0.31 |
ENST00000339023.4
ENST00000354664.4 |
TBC1D3
|
TBC1 domain family, member 3 |
| chr11_+_57365150 | 0.30 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr10_-_106240032 | 0.29 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr6_+_56820018 | 0.29 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr11_-_73882029 | 0.29 |
ENST00000539061.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr5_+_7869217 | 0.29 |
ENST00000264668.2
ENST00000514220.1 ENST00000341013.6 ENST00000440940.2 ENST00000502550.1 ENST00000506877.1 |
MTRR
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr19_+_56652686 | 0.29 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr17_-_34808047 | 0.29 |
ENST00000592614.1
ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G
TBC1D3H
|
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr3_-_112218205 | 0.29 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr16_-_67260691 | 0.28 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr8_+_95653302 | 0.28 |
ENST00000423620.2
ENST00000433389.2 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr10_-_76995769 | 0.28 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr12_-_25348007 | 0.28 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr14_-_101295407 | 0.28 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr17_-_79359021 | 0.28 |
ENST00000572301.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr3_-_3151664 | 0.28 |
ENST00000256452.3
ENST00000311981.8 ENST00000430514.2 ENST00000456302.1 |
IL5RA
|
interleukin 5 receptor, alpha |
| chr10_+_99627889 | 0.27 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr17_-_34807272 | 0.27 |
ENST00000535592.1
ENST00000394453.1 |
TBC1D3G
|
TBC1 domain family, member 3G |
| chr7_-_107770794 | 0.27 |
ENST00000205386.4
ENST00000418464.1 ENST00000388781.3 ENST00000388780.3 ENST00000414450.2 |
LAMB4
|
laminin, beta 4 |
| chr17_-_36347835 | 0.27 |
ENST00000519532.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr1_+_52082751 | 0.27 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VDR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 1.0 | 7.0 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.9 | 2.7 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.8 | 2.4 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.8 | 2.4 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.7 | 4.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.7 | 2.7 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) phenylpropanoid catabolic process(GO:0046271) |
| 0.7 | 2.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.6 | 2.5 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.6 | 3.5 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.5 | 2.6 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.3 | 1.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.3 | 1.0 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 1.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 6.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 2.9 | GO:0051918 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 0.9 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 0.8 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 | 1.6 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 0.9 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.2 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.5 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 1.9 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.4 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 4.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.6 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 2.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 1.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.4 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.3 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.3 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 1.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.9 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.7 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 2.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.7 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.4 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 0.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.3 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.5 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.3 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.4 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.5 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0071264 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.0 | 0.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.4 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.9 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.6 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 1.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 2.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.6 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 1.5 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.3 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.4 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0097181 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.9 | 3.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 4.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 3.0 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 2.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 0.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 1.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 5.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.9 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 1.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.8 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 3.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.2 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 3.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.9 | 2.7 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.8 | 2.4 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.8 | 5.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.6 | 1.9 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.6 | 3.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.5 | 2.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.5 | 2.7 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.4 | 4.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 1.6 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.3 | 1.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.3 | 2.4 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.3 | 1.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 0.6 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.2 | 1.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 4.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 4.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.7 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.9 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.6 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 0.8 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.5 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 2.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 1.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.4 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.5 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.4 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 1.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.5 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 1.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 2.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.3 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.6 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 2.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 5.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 6.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 2.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 7.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |