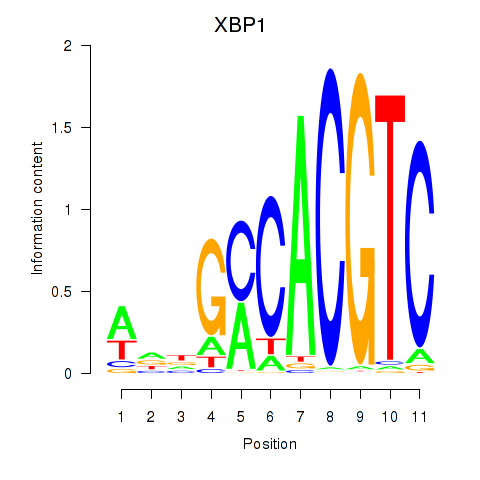
|
chr7_+_94023873
Show fit
|
6.12 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr19_-_55895966
Show fit
|
3.84 |
ENST00000444469.3
|
TMEM238
|
transmembrane protein 238
|
|
chr12_+_56624436
Show fit
|
3.83 |
ENST00000266980.4
ENST00000437277.1
|
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5
|
|
chr10_+_89419370
Show fit
|
3.78 |
ENST00000361175.4
ENST00000456849.1
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr22_+_38864041
Show fit
|
3.62 |
ENST00000216014.4
ENST00000409006.3
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr3_-_132378897
Show fit
|
2.99 |
ENST00000545291.1
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11
|
|
chr3_-_132378919
Show fit
|
2.95 |
ENST00000355458.3
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11
|
|
chr11_+_69455855
Show fit
|
2.51 |
ENST00000227507.2
ENST00000536559.1
|
CCND1
|
cyclin D1
|
|
chr1_+_21835858
Show fit
|
2.41 |
ENST00000539907.1
ENST00000540617.1
ENST00000374840.3
|
ALPL
|
alkaline phosphatase, liver/bone/kidney
|
|
chr7_-_27183263
Show fit
|
2.37 |
ENST00000222726.3
|
HOXA5
|
homeobox A5
|
|
chr19_+_35773242
Show fit
|
2.30 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chr3_-_57583052
Show fit
|
2.19 |
ENST00000496292.1
ENST00000489843.1
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr4_-_119757239
Show fit
|
2.16 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D
|
|
chr6_+_132129151
Show fit
|
2.14 |
ENST00000360971.2
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr1_-_24126051
Show fit
|
2.13 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr10_+_102106829
Show fit
|
2.11 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr7_+_16793160
Show fit
|
1.99 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13
|
|
chr11_+_17298255
Show fit
|
1.90 |
ENST00000531172.1
ENST00000533738.2
ENST00000323688.6
|
NUCB2
|
nucleobindin 2
|
|
chr3_-_57583185
Show fit
|
1.89 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr11_+_17298297
Show fit
|
1.87 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2
|
|
chr3_+_171758344
Show fit
|
1.86 |
ENST00000336824.4
ENST00000423424.1
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr3_-_57583130
Show fit
|
1.86 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr3_-_128369643
Show fit
|
1.82 |
ENST00000296255.3
|
RPN1
|
ribophorin I
|
|
chr4_+_141445333
Show fit
|
1.80 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2
|
|
chrX_+_153060090
Show fit
|
1.79 |
ENST00000370086.3
ENST00000370085.3
|
SSR4
|
signal sequence receptor, delta
|
|
chr12_-_99038732
Show fit
|
1.77 |
ENST00000393042.3
ENST00000420861.1
ENST00000299157.4
ENST00000342502.2
|
IKBIP
|
IKBKB interacting protein
|
|
chr1_-_24126023
Show fit
|
1.77 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr19_+_49468558
Show fit
|
1.70 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide
|
|
chr4_-_119757322
Show fit
|
1.63 |
ENST00000379735.5
|
SEC24D
|
SEC24 family member D
|
|
chrX_+_153059608
Show fit
|
1.61 |
ENST00000370087.1
|
SSR4
|
signal sequence receptor, delta
|
|
chr11_-_118972575
Show fit
|
1.61 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase)
|
|
chr5_-_2751762
Show fit
|
1.61 |
ENST00000302057.5
ENST00000382611.6
|
IRX2
|
iroquois homeobox 2
|
|
chr8_-_38126675
Show fit
|
1.59 |
ENST00000531823.1
ENST00000534339.1
ENST00000524616.1
ENST00000422581.2
ENST00000424479.2
ENST00000419686.2
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chr11_+_17298543
Show fit
|
1.57 |
ENST00000533926.1
|
NUCB2
|
nucleobindin 2
|
|
chr6_+_33168189
Show fit
|
1.55 |
ENST00000444757.1
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr9_+_124461603
Show fit
|
1.52 |
ENST00000373782.3
|
DAB2IP
|
DAB2 interacting protein
|
|
chr5_+_9546306
Show fit
|
1.51 |
ENST00000508179.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding)
|
|
chr20_-_17662705
Show fit
|
1.51 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1
|
|
chr12_+_107349497
Show fit
|
1.50 |
ENST00000548125.1
ENST00000280756.4
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr6_+_33168597
Show fit
|
1.47 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr3_+_127771212
Show fit
|
1.46 |
ENST00000243253.3
ENST00000481210.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr16_+_2083265
Show fit
|
1.46 |
ENST00000565855.1
ENST00000566198.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2
|
|
chr11_+_17298522
Show fit
|
1.42 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2
|
|
chr9_+_114393581
Show fit
|
1.38 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25
|
|
chr5_+_9546376
Show fit
|
1.35 |
ENST00000509788.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding)
|
|
chr5_-_121413974
Show fit
|
1.35 |
ENST00000231004.4
|
LOX
|
lysyl oxidase
|
|
chr6_+_33168637
Show fit
|
1.32 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr11_+_17298342
Show fit
|
1.31 |
ENST00000530964.1
|
NUCB2
|
nucleobindin 2
|
|
chr14_-_24664776
Show fit
|
1.28 |
ENST00000530468.1
ENST00000528010.1
ENST00000396854.4
ENST00000524835.1
ENST00000261789.4
ENST00000525592.1
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr12_-_106641728
Show fit
|
1.26 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr20_-_17662878
Show fit
|
1.25 |
ENST00000377813.1
ENST00000377807.2
ENST00000360807.4
ENST00000398782.2
|
RRBP1
|
ribosome binding protein 1
|
|
chr11_-_118927816
Show fit
|
1.23 |
ENST00000534233.1
ENST00000532752.1
ENST00000525859.1
ENST00000404233.3
ENST00000532421.1
ENST00000543287.1
ENST00000527310.2
ENST00000529972.1
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr3_+_133524459
Show fit
|
1.22 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit
|
|
chr19_-_4670345
Show fit
|
1.21 |
ENST00000599630.1
ENST00000262947.3
|
C19orf10
|
chromosome 19 open reading frame 10
|
|
chr14_+_100259666
Show fit
|
1.20 |
ENST00000262233.6
ENST00000334192.4
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr14_+_39736582
Show fit
|
1.20 |
ENST00000556148.1
ENST00000348007.3
|
CTAGE5
|
CTAGE family, member 5
|
|
chr7_-_6523755
Show fit
|
1.18 |
ENST00000436575.1
ENST00000258739.4
|
DAGLB
KDELR2
|
diacylglycerol lipase, beta
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr3_+_132379154
Show fit
|
1.16 |
ENST00000468022.1
ENST00000473651.1
ENST00000494238.2
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr7_+_97840739
Show fit
|
1.16 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15
|
|
chr14_-_24664540
Show fit
|
1.13 |
ENST00000530563.1
ENST00000528895.1
ENST00000528669.1
ENST00000532632.1
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr2_-_69614373
Show fit
|
1.13 |
ENST00000361060.5
ENST00000357308.4
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr11_-_68039364
Show fit
|
1.12 |
ENST00000533310.1
ENST00000304271.6
ENST00000527280.1
|
C11orf24
|
chromosome 11 open reading frame 24
|
|
chr8_-_124054484
Show fit
|
1.07 |
ENST00000419562.2
|
DERL1
|
derlin 1
|
|
chr7_-_6523688
Show fit
|
1.06 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr8_-_124054362
Show fit
|
1.04 |
ENST00000405944.3
|
DERL1
|
derlin 1
|
|
chr4_+_128982416
Show fit
|
1.02 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr9_-_112260531
Show fit
|
0.99 |
ENST00000374541.2
ENST00000262539.3
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr14_+_100259712
Show fit
|
0.99 |
ENST00000556714.1
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr13_+_46039037
Show fit
|
0.99 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr9_+_139377947
Show fit
|
0.98 |
ENST00000354376.1
|
C9orf163
|
chromosome 9 open reading frame 163
|
|
chr1_-_119683251
Show fit
|
0.98 |
ENST00000369426.5
ENST00000235521.4
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial
|
|
chr1_+_26758849
Show fit
|
0.98 |
ENST00000533087.1
ENST00000531312.1
ENST00000525165.1
ENST00000525326.1
ENST00000525546.1
ENST00000436153.2
ENST00000530781.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr12_+_56862301
Show fit
|
0.97 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4
|
|
chr11_-_66056596
Show fit
|
0.96 |
ENST00000471387.2
ENST00000359461.6
ENST00000376901.4
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr5_+_176730769
Show fit
|
0.94 |
ENST00000303204.4
ENST00000503216.1
|
PRELID1
|
PRELI domain containing 1
|
|
chr11_-_118927880
Show fit
|
0.94 |
ENST00000527038.2
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr1_+_101361782
Show fit
|
0.93 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr9_+_101984577
Show fit
|
0.92 |
ENST00000223641.4
|
SEC61B
|
Sec61 beta subunit
|
|
chr8_-_124054587
Show fit
|
0.91 |
ENST00000259512.4
|
DERL1
|
derlin 1
|
|
chr2_-_182521823
Show fit
|
0.91 |
ENST00000410087.3
ENST00000409440.3
|
CERKL
|
ceramide kinase-like
|
|
chr1_-_87379785
Show fit
|
0.90 |
ENST00000401030.3
ENST00000370554.1
|
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA.
|
|
chr3_-_150264272
Show fit
|
0.89 |
ENST00000491660.1
ENST00000487153.1
ENST00000239944.2
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr7_-_128045984
Show fit
|
0.89 |
ENST00000470772.1
ENST00000480861.1
ENST00000496200.1
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr1_+_26758790
Show fit
|
0.89 |
ENST00000427245.2
ENST00000525682.2
ENST00000236342.7
ENST00000526219.1
ENST00000374185.3
ENST00000360009.2
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr1_-_119682812
Show fit
|
0.88 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial
|
|
chr1_-_87380002
Show fit
|
0.88 |
ENST00000331835.5
|
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA.
|
|
chr3_-_10362725
Show fit
|
0.87 |
ENST00000397109.3
ENST00000428626.1
ENST00000445064.1
ENST00000431352.1
ENST00000397117.1
ENST00000337354.4
ENST00000383801.2
ENST00000432213.1
ENST00000350697.3
|
SEC13
|
SEC13 homolog (S. cerevisiae)
|
|
chr11_-_118927561
Show fit
|
0.87 |
ENST00000530473.1
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr17_+_46048497
Show fit
|
0.87 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|
|
chr19_-_29704448
Show fit
|
0.87 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
|
|
chr2_-_38604398
Show fit
|
0.85 |
ENST00000443098.1
ENST00000449130.1
ENST00000378954.4
ENST00000539122.1
ENST00000419554.2
ENST00000451483.1
ENST00000406122.1
|
ATL2
|
atlastin GTPase 2
|
|
chr1_-_45988542
Show fit
|
0.85 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1
|
|
chr17_+_46048376
Show fit
|
0.85 |
ENST00000338399.4
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|
|
chr11_-_66056478
Show fit
|
0.85 |
ENST00000431556.2
ENST00000528575.1
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr10_+_104678102
Show fit
|
0.84 |
ENST00000433628.2
|
CNNM2
|
cyclin M2
|
|
chr6_-_109702885
Show fit
|
0.84 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin
|
|
chr18_-_47018869
Show fit
|
0.83 |
ENST00000583036.1
ENST00000580261.1
|
RPL17
|
ribosomal protein L17
|
|
chr2_+_27255806
Show fit
|
0.82 |
ENST00000238788.9
ENST00000404032.3
|
TMEM214
|
transmembrane protein 214
|
|
chr10_+_18948311
Show fit
|
0.81 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr2_-_88927092
Show fit
|
0.80 |
ENST00000303236.3
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr11_-_118927735
Show fit
|
0.79 |
ENST00000526656.1
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr11_-_207221
Show fit
|
0.78 |
ENST00000486280.1
ENST00000332865.6
ENST00000529614.2
ENST00000325147.9
ENST00000410108.1
ENST00000382762.3
|
BET1L
|
Bet1 golgi vesicular membrane trafficking protein-like
|
|
chr4_+_141445311
Show fit
|
0.77 |
ENST00000323570.3
ENST00000511887.2
|
ELMOD2
|
ELMO/CED-12 domain containing 2
|
|
chr6_+_63921351
Show fit
|
0.77 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C
|
|
chr22_+_46476192
Show fit
|
0.76 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763
|
|
chr3_+_105086056
Show fit
|
0.76 |
ENST00000472644.2
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr17_+_46048471
Show fit
|
0.76 |
ENST00000578018.1
ENST00000579175.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|
|
chr2_-_220408430
Show fit
|
0.76 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor
|
|
chr3_+_132378741
Show fit
|
0.75 |
ENST00000493720.2
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr11_-_62414070
Show fit
|
0.74 |
ENST00000540933.1
ENST00000346178.4
ENST00000356638.3
ENST00000534779.1
ENST00000525994.1
|
GANAB
|
glucosidase, alpha; neutral AB
|
|
chr1_+_6845497
Show fit
|
0.74 |
ENST00000473578.1
ENST00000557126.1
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr5_+_154237778
Show fit
|
0.74 |
ENST00000523698.1
ENST00000517876.1
ENST00000520472.1
|
CNOT8
|
CCR4-NOT transcription complex, subunit 8
|
|
chr19_+_39421556
Show fit
|
0.73 |
ENST00000407800.2
ENST00000402029.3
|
MRPS12
|
mitochondrial ribosomal protein S12
|
|
chr14_+_50087468
Show fit
|
0.73 |
ENST00000305386.2
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr15_+_79603404
Show fit
|
0.73 |
ENST00000299705.5
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr10_-_22292675
Show fit
|
0.72 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr19_+_1491144
Show fit
|
0.72 |
ENST00000233596.3
|
REEP6
|
receptor accessory protein 6
|
|
chr6_-_83902933
Show fit
|
0.72 |
ENST00000512866.1
ENST00000510258.1
ENST00000503094.1
ENST00000283977.4
ENST00000513973.1
ENST00000508748.1
|
PGM3
|
phosphoglucomutase 3
|
|
chr9_+_114393634
Show fit
|
0.71 |
ENST00000556107.1
ENST00000374294.3
|
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25
DNAJC25-GNG10 readthrough
|
|
chr1_-_155145721
Show fit
|
0.71 |
ENST00000295682.4
|
KRTCAP2
|
keratinocyte associated protein 2
|
|
chr4_+_128982490
Show fit
|
0.69 |
ENST00000394288.3
ENST00000432347.2
ENST00000264584.5
ENST00000441387.1
ENST00000427266.1
ENST00000354456.3
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr5_+_126853301
Show fit
|
0.69 |
ENST00000296666.8
ENST00000442138.2
ENST00000512635.2
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr18_-_47018897
Show fit
|
0.69 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17
|
|
chr14_+_39736299
Show fit
|
0.68 |
ENST00000341502.5
ENST00000396158.2
ENST00000280083.3
|
CTAGE5
|
CTAGE family, member 5
|
|
chr6_-_34664612
Show fit
|
0.68 |
ENST00000374023.3
ENST00000374026.3
|
C6orf106
|
chromosome 6 open reading frame 106
|
|
chr1_-_154946792
Show fit
|
0.67 |
ENST00000412170.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr10_+_112257596
Show fit
|
0.66 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr11_-_59383617
Show fit
|
0.66 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein
|
|
chr5_+_154238042
Show fit
|
0.65 |
ENST00000519211.1
ENST00000522458.1
ENST00000519903.1
ENST00000521450.1
ENST00000403027.2
|
CNOT8
|
CCR4-NOT transcription complex, subunit 8
|
|
chr8_-_81083890
Show fit
|
0.62 |
ENST00000518937.1
|
TPD52
|
tumor protein D52
|
|
chr7_+_128379346
Show fit
|
0.60 |
ENST00000535011.2
ENST00000542996.2
ENST00000535623.1
ENST00000538546.1
ENST00000249364.4
ENST00000449187.2
|
CALU
|
calumenin
|
|
chr2_-_220408260
Show fit
|
0.60 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor
|
|
chr4_+_108746282
Show fit
|
0.59 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr4_+_128982430
Show fit
|
0.59 |
ENST00000512292.1
ENST00000508819.1
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr17_+_8339189
Show fit
|
0.59 |
ENST00000585098.1
ENST00000380025.4
ENST00000402554.3
ENST00000584866.1
ENST00000582490.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1
|
|
chr2_+_241508039
Show fit
|
0.58 |
ENST00000270357.4
|
RNPEPL1
|
arginyl aminopeptidase (aminopeptidase B)-like 1
|
|
chr7_+_127292234
Show fit
|
0.58 |
ENST00000354725.3
|
SND1
|
staphylococcal nuclease and tudor domain containing 1
|
|
chr19_+_42388437
Show fit
|
0.58 |
ENST00000378152.4
ENST00000337665.4
|
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr1_+_110527308
Show fit
|
0.57 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr20_-_62199427
Show fit
|
0.57 |
ENST00000427522.2
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator
|
|
chr6_+_41606176
Show fit
|
0.57 |
ENST00000441667.1
ENST00000230321.6
ENST00000373050.4
ENST00000446650.1
ENST00000435476.1
|
MDFI
|
MyoD family inhibitor
|
|
chr7_+_128379449
Show fit
|
0.56 |
ENST00000479257.1
|
CALU
|
calumenin
|
|
chrX_-_41782249
Show fit
|
0.56 |
ENST00000442742.2
ENST00000421587.2
ENST00000378166.4
ENST00000318588.9
ENST00000361962.4
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr18_-_47017956
Show fit
|
0.55 |
ENST00000584895.1
ENST00000332968.6
ENST00000580210.1
ENST00000579408.1
|
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough
ribosomal protein L17
|
|
chr5_-_115177496
Show fit
|
0.55 |
ENST00000274459.4
ENST00000509910.1
|
ATG12
|
autophagy related 12
|
|
chr11_-_62599505
Show fit
|
0.55 |
ENST00000377897.4
ENST00000394690.1
ENST00000541317.1
ENST00000294179.3
|
STX5
|
syntaxin 5
|
|
chr9_-_35072585
Show fit
|
0.54 |
ENST00000448530.1
|
VCP
|
valosin containing protein
|
|
chrX_+_153627231
Show fit
|
0.54 |
ENST00000406022.2
|
RPL10
|
ribosomal protein L10
|
|
chr19_+_1026566
Show fit
|
0.54 |
ENST00000348419.3
ENST00000565096.2
ENST00000562958.2
ENST00000562075.2
ENST00000607102.1
|
CNN2
|
calponin 2
|
|
chr19_+_33182823
Show fit
|
0.54 |
ENST00000397061.3
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19
|
|
chr5_+_119799927
Show fit
|
0.54 |
ENST00000407149.2
ENST00000379551.2
|
PRR16
|
proline rich 16
|
|
chr19_+_7701985
Show fit
|
0.53 |
ENST00000595950.1
ENST00000441779.2
ENST00000221283.5
ENST00000414284.2
|
STXBP2
|
syntaxin binding protein 2
|
|
chr10_-_30348439
Show fit
|
0.52 |
ENST00000375377.1
|
KIAA1462
|
KIAA1462
|
|
chr2_-_20251744
Show fit
|
0.52 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr17_-_40575535
Show fit
|
0.51 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor
|
|
chr3_-_139108475
Show fit
|
0.50 |
ENST00000515006.1
ENST00000513274.1
ENST00000514508.1
ENST00000507777.1
ENST00000512153.1
ENST00000333188.5
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime)
|
|
chr5_-_9546180
Show fit
|
0.50 |
ENST00000382496.5
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr10_-_128077024
Show fit
|
0.50 |
ENST00000368679.4
ENST00000368676.4
ENST00000448723.1
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr1_+_207669573
Show fit
|
0.49 |
ENST00000400960.2
ENST00000534202.1
|
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group)
|
|
chr10_+_104678032
Show fit
|
0.48 |
ENST00000369878.4
ENST00000369875.3
|
CNNM2
|
cyclin M2
|
|
chr6_-_7313381
Show fit
|
0.48 |
ENST00000489567.1
ENST00000479365.1
ENST00000462112.1
ENST00000397511.2
ENST00000534851.1
ENST00000474597.1
ENST00000244763.4
|
SSR1
|
signal sequence receptor, alpha
|
|
chr15_-_75165651
Show fit
|
0.48 |
ENST00000562363.1
ENST00000564529.1
ENST00000268099.9
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr19_-_19030157
Show fit
|
0.46 |
ENST00000349893.4
ENST00000351079.4
ENST00000600932.1
ENST00000262812.4
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr6_+_7108210
Show fit
|
0.44 |
ENST00000467782.1
ENST00000334984.6
ENST00000349384.6
|
RREB1
|
ras responsive element binding protein 1
|
|
chr15_-_60690932
Show fit
|
0.44 |
ENST00000559818.1
|
ANXA2
|
annexin A2
|
|
chr22_+_45148432
Show fit
|
0.44 |
ENST00000389774.2
ENST00000396119.2
ENST00000336963.4
ENST00000356099.6
ENST00000412433.1
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr19_-_10426663
Show fit
|
0.43 |
ENST00000541276.1
ENST00000393708.3
ENST00000494368.1
|
FDX1L
|
ferredoxin 1-like
|
|
chr17_+_8339164
Show fit
|
0.43 |
ENST00000582665.1
ENST00000334527.7
ENST00000299734.7
|
NDEL1
|
nudE neurodevelopment protein 1-like 1
|
|
chr5_-_60458179
Show fit
|
0.43 |
ENST00000507416.1
ENST00000339020.3
|
SMIM15
|
small integral membrane protein 15
|
|
chrX_-_41782683
Show fit
|
0.43 |
ENST00000378163.1
ENST00000378154.1
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr12_+_7053228
Show fit
|
0.42 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr11_+_47430133
Show fit
|
0.41 |
ENST00000531974.1
ENST00000531419.1
ENST00000531865.1
ENST00000362021.4
ENST00000354884.4
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr5_+_154238096
Show fit
|
0.41 |
ENST00000517568.1
ENST00000524105.1
ENST00000285896.6
|
CNOT8
|
CCR4-NOT transcription complex, subunit 8
|
|
chr19_+_16187816
Show fit
|
0.40 |
ENST00000588410.1
|
TPM4
|
tropomyosin 4
|
|
chr13_-_45915221
Show fit
|
0.40 |
ENST00000309246.5
ENST00000379060.4
ENST00000379055.1
ENST00000527226.1
ENST00000379056.1
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr17_+_73106035
Show fit
|
0.40 |
ENST00000581078.1
ENST00000582136.1
ENST00000245543.1
|
ARMC7
|
armadillo repeat containing 7
|
|
chr20_-_62493217
Show fit
|
0.40 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135
|
|
chr3_-_105587879
Show fit
|
0.39 |
ENST00000264122.4
ENST00000403724.1
ENST00000405772.1
|
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase
|
|
chr5_+_154238149
Show fit
|
0.39 |
ENST00000519430.1
ENST00000520671.1
ENST00000521583.1
ENST00000518028.1
ENST00000519404.1
ENST00000519394.1
ENST00000518775.1
|
CNOT8
|
CCR4-NOT transcription complex, subunit 8
|
|
chr3_+_33155525
Show fit
|
0.39 |
ENST00000449224.1
|
CRTAP
|
cartilage associated protein
|
|
chr12_-_56123444
Show fit
|
0.39 |
ENST00000546457.1
ENST00000549117.1
|
CD63
|
CD63 molecule
|
|
chr4_+_56262115
Show fit
|
0.38 |
ENST00000506198.1
ENST00000381334.5
ENST00000542052.1
|
TMEM165
|
transmembrane protein 165
|
|
chr1_+_1567474
Show fit
|
0.38 |
ENST00000356026.5
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr15_+_79603479
Show fit
|
0.37 |
ENST00000424155.2
ENST00000536821.1
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr14_-_50087312
Show fit
|
0.37 |
ENST00000298289.6
|
RPL36AL
|
ribosomal protein L36a-like
|
|
chr19_-_39523165
Show fit
|
0.37 |
ENST00000509137.2
ENST00000292853.4
|
FBXO27
|
F-box protein 27
|
|
chr1_+_6845578
Show fit
|
0.36 |
ENST00000467404.2
ENST00000439411.2
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr3_-_156272872
Show fit
|
0.35 |
ENST00000476217.1
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma)
|
|
chr6_-_108279369
Show fit
|
0.35 |
ENST00000369002.4
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr17_+_72199721
Show fit
|
0.35 |
ENST00000439590.2
ENST00000311111.6
ENST00000584577.1
ENST00000534490.1
ENST00000528433.2
ENST00000533498.1
|
RPL38
|
ribosomal protein L38
|
|
chr15_-_60690163
Show fit
|
0.34 |
ENST00000558998.1
ENST00000560165.1
ENST00000557986.1
ENST00000559780.1
ENST00000559467.1
ENST00000559956.1
ENST00000332680.4
ENST00000396024.3
ENST00000421017.2
ENST00000560466.1
ENST00000558132.1
ENST00000559113.1
ENST00000557906.1
ENST00000558558.1
ENST00000560468.1
ENST00000559370.1
ENST00000558169.1
ENST00000559725.1
ENST00000558985.1
ENST00000451270.2
|
ANXA2
|
annexin A2
|
|
chr11_+_61595572
Show fit
|
0.33 |
ENST00000517312.1
|
FADS2
|
fatty acid desaturase 2
|
|
chr6_+_116692102
Show fit
|
0.33 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase
|
|
chr16_+_56965960
Show fit
|
0.33 |
ENST00000439977.2
ENST00000344114.4
ENST00000300302.5
ENST00000379792.2
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr7_+_6522922
Show fit
|
0.32 |
ENST00000601673.1
|
FLJ20306
|
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein
|
|
chr2_-_242254595
Show fit
|
0.31 |
ENST00000441124.1
ENST00000391976.2
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr17_+_8339340
Show fit
|
0.31 |
ENST00000580012.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1
|
|
chr19_-_8373173
Show fit
|
0.31 |
ENST00000537716.2
ENST00000301458.5
|
CD320
|
CD320 molecule
|
|
chr1_+_207669613
Show fit
|
0.30 |
ENST00000367049.4
ENST00000529814.1
|
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group)
|
|
chr1_+_27669719
Show fit
|
0.30 |
ENST00000473280.1
|
SYTL1
|
synaptotagmin-like 1
|