Project
Illumina Body Map 2
Navigation
Downloads
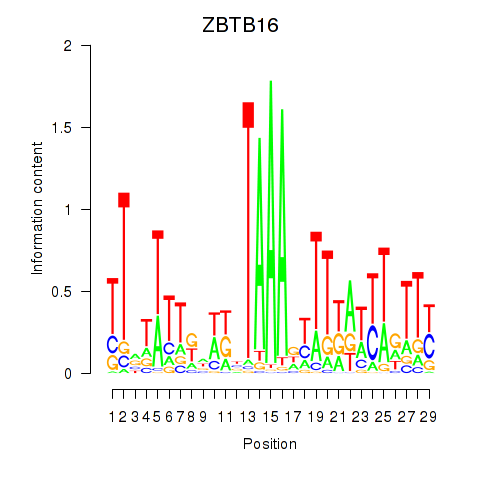
Results for ZBTB16
Z-value: 0.96
Transcription factors associated with ZBTB16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB16
|
ENSG00000109906.9 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB16 | hg19_v2_chr11_+_113930291_113930339 | -0.71 | 5.4e-06 | Click! |
Activity profile of ZBTB16 motif
Sorted Z-values of ZBTB16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_48082192 | 4.53 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr6_-_49712123 | 4.08 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_-_49712091 | 3.68 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr12_-_53601055 | 3.08 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr12_-_53601000 | 2.90 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr5_-_55412774 | 2.74 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr20_+_38633006 | 2.57 |
ENST00000432633.1
ENST00000445606.1 |
RP11-101E14.2
|
RP11-101E14.2 |
| chr4_-_38784572 | 2.53 |
ENST00000308973.4
ENST00000361424.2 |
TLR10
|
toll-like receptor 10 |
| chr1_+_207277632 | 2.51 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr4_-_38784592 | 2.48 |
ENST00000502321.1
|
TLR10
|
toll-like receptor 10 |
| chr7_-_38289173 | 2.48 |
ENST00000436911.2
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr1_+_198608292 | 2.29 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr1_+_198608146 | 2.13 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_89901292 | 2.11 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr6_-_49712147 | 2.10 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr3_-_120365866 | 2.05 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr14_+_22670455 | 1.91 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr15_-_55562479 | 1.83 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr5_+_35856951 | 1.78 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr14_+_22458631 | 1.76 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr4_+_74347400 | 1.75 |
ENST00000226355.3
|
AFM
|
afamin |
| chr2_+_204801471 | 1.73 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr19_+_14693888 | 1.73 |
ENST00000547437.1
ENST00000397439.2 ENST00000417570.1 |
CLEC17A
|
C-type lectin domain family 17, member A |
| chr12_-_120765565 | 1.72 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr14_+_22564294 | 1.72 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chr15_-_55562582 | 1.69 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_-_89310012 | 1.68 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr10_+_90521163 | 1.66 |
ENST00000404459.1
|
LIPN
|
lipase, family member N |
| chr12_+_10460549 | 1.66 |
ENST00000543420.1
ENST00000543777.1 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr6_-_49712072 | 1.64 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr7_-_36764142 | 1.62 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr6_-_44233361 | 1.60 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr19_+_45690646 | 1.54 |
ENST00000591569.1
|
AC006126.3
|
Uncharacterized protein |
| chr4_+_36283213 | 1.51 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr3_+_46395219 | 1.50 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr15_-_55562451 | 1.50 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_+_42041860 | 1.48 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr12_+_21284118 | 1.48 |
ENST00000256958.2
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1 |
| chr7_-_36764004 | 1.46 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr7_-_36764062 | 1.44 |
ENST00000435386.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr9_-_116837249 | 1.40 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr7_+_76090993 | 1.38 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr4_+_74606223 | 1.32 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr4_+_154622652 | 1.31 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr19_-_55150343 | 1.28 |
ENST00000456337.1
|
AC009892.10
|
Uncharacterized protein |
| chr15_-_54051831 | 1.28 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr14_+_22788560 | 1.25 |
ENST00000390468.1
|
TRAV41
|
T cell receptor alpha variable 41 |
| chr15_-_54025300 | 1.23 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr10_+_7745232 | 1.19 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_+_200011711 | 1.16 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr10_+_7745303 | 1.16 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chrX_+_12885183 | 1.13 |
ENST00000380659.3
|
TLR7
|
toll-like receptor 7 |
| chr19_+_42041702 | 1.09 |
ENST00000487420.2
|
AC006129.4
|
AC006129.4 |
| chr4_-_39033963 | 1.09 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr17_-_46623441 | 1.08 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr12_+_4385230 | 1.06 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr1_-_100231349 | 1.03 |
ENST00000287474.5
ENST00000414213.1 |
FRRS1
|
ferric-chelate reductase 1 |
| chr16_+_81812863 | 1.02 |
ENST00000359376.3
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr12_+_20968608 | 1.00 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr10_-_10504285 | 0.98 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr2_+_90229045 | 0.97 |
ENST00000390278.2
|
IGKV1D-42
|
immunoglobulin kappa variable 1D-42 (non-functional) |
| chr3_-_4927447 | 0.96 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr2_+_102618428 | 0.95 |
ENST00000457817.1
|
IL1R2
|
interleukin 1 receptor, type II |
| chr1_+_241695670 | 0.93 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr1_+_196857144 | 0.92 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr2_+_181988620 | 0.90 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr19_+_42041537 | 0.88 |
ENST00000597199.1
|
AC006129.4
|
AC006129.4 |
| chr6_+_27925019 | 0.86 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr6_+_106988986 | 0.84 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr6_+_32812568 | 0.84 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_-_190448118 | 0.84 |
ENST00000440626.1
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr12_-_68696652 | 0.84 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr12_-_10007448 | 0.84 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_-_113772835 | 0.83 |
ENST00000552014.1
ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr9_-_71161505 | 0.82 |
ENST00000446290.1
|
RP11-274B18.2
|
RP11-274B18.2 |
| chr15_-_50406680 | 0.81 |
ENST00000559829.1
|
ATP8B4
|
ATPase, class I, type 8B, member 4 |
| chr2_+_192543694 | 0.79 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr2_-_61245363 | 0.79 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr10_+_8096769 | 0.78 |
ENST00000346208.3
|
GATA3
|
GATA binding protein 3 |
| chr14_-_45722360 | 0.77 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr1_+_174769006 | 0.76 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr5_+_96212185 | 0.76 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr17_+_40925454 | 0.75 |
ENST00000253794.2
ENST00000590339.1 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr12_+_10460417 | 0.75 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr14_+_77228532 | 0.73 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr6_+_42749759 | 0.71 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr2_-_32490859 | 0.69 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chrX_+_115301975 | 0.68 |
ENST00000371906.4
|
AGTR2
|
angiotensin II receptor, type 2 |
| chr6_+_136172820 | 0.63 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr11_-_327537 | 0.63 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr19_+_3762645 | 0.61 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr10_+_48189612 | 0.61 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr3_-_146262637 | 0.60 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr10_+_8096631 | 0.59 |
ENST00000379328.3
|
GATA3
|
GATA binding protein 3 |
| chr7_-_107770794 | 0.59 |
ENST00000205386.4
ENST00000418464.1 ENST00000388781.3 ENST00000388780.3 ENST00000414450.2 |
LAMB4
|
laminin, beta 4 |
| chr5_+_180682720 | 0.59 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr11_-_104972158 | 0.58 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr8_-_30585439 | 0.58 |
ENST00000221130.5
|
GSR
|
glutathione reductase |
| chr16_+_19183671 | 0.57 |
ENST00000562711.2
|
SYT17
|
synaptotagmin XVII |
| chr8_-_8318847 | 0.56 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr17_-_295730 | 0.56 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr20_-_54580523 | 0.56 |
ENST00000064571.2
|
CBLN4
|
cerebellin 4 precursor |
| chr3_-_93692781 | 0.56 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr10_-_47239738 | 0.55 |
ENST00000413193.2
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr19_+_3762703 | 0.55 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr22_-_30901637 | 0.55 |
ENST00000381982.3
ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4
|
SEC14-like 4 (S. cerevisiae) |
| chr8_+_66619277 | 0.55 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr2_-_104496621 | 0.54 |
ENST00000455716.1
|
AC013727.1
|
AC013727.1 |
| chr4_-_88244010 | 0.54 |
ENST00000302219.6
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr19_-_56343353 | 0.54 |
ENST00000592953.1
ENST00000589093.1 |
NLRP11
|
NLR family, pyrin domain containing 11 |
| chr14_+_22976633 | 0.53 |
ENST00000390503.1
|
TRAJ34
|
T cell receptor alpha joining 34 |
| chr5_-_54988448 | 0.53 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr4_+_71263599 | 0.53 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr4_-_76957214 | 0.52 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr3_-_146262488 | 0.51 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr22_+_30821732 | 0.51 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr11_-_111649074 | 0.50 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr10_-_74283694 | 0.48 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr6_+_26273144 | 0.47 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr4_-_88244049 | 0.46 |
ENST00000328546.4
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_+_159848807 | 0.46 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr22_+_30821784 | 0.46 |
ENST00000407550.3
|
MTFP1
|
mitochondrial fission process 1 |
| chr5_-_41071392 | 0.44 |
ENST00000399564.4
|
MROH2B
|
maestro heat-like repeat family member 2B |
| chr15_+_75628232 | 0.44 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr7_+_120591170 | 0.44 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr16_+_68071816 | 0.44 |
ENST00000562246.1
|
DUS2
|
dihydrouridine synthase 2 |
| chr8_-_48651648 | 0.42 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr13_-_101240985 | 0.42 |
ENST00000471912.1
|
GGACT
|
gamma-glutamylamine cyclotransferase |
| chr17_-_37607497 | 0.41 |
ENST00000394287.3
ENST00000300651.6 |
MED1
|
mediator complex subunit 1 |
| chr11_+_43380459 | 0.40 |
ENST00000299240.6
ENST00000039989.4 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr9_+_79074068 | 0.39 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr19_+_45281118 | 0.39 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr19_-_23869999 | 0.37 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr6_+_25963020 | 0.36 |
ENST00000357085.3
|
TRIM38
|
tripartite motif containing 38 |
| chr2_+_181988560 | 0.35 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr16_+_22501658 | 0.35 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr13_+_77564795 | 0.35 |
ENST00000377453.3
|
CLN5
|
ceroid-lipofuscinosis, neuronal 5 |
| chrX_-_2882296 | 0.35 |
ENST00000438544.1
ENST00000381134.3 ENST00000545496.1 |
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1) |
| chr8_+_104831472 | 0.35 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr12_+_105380073 | 0.35 |
ENST00000552951.1
ENST00000280749.5 |
C12orf45
|
chromosome 12 open reading frame 45 |
| chr14_+_39703112 | 0.35 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr15_+_81391740 | 0.34 |
ENST00000561216.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr11_-_66445219 | 0.34 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
| chr11_-_111649015 | 0.34 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr12_+_133758115 | 0.34 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr1_+_247712383 | 0.34 |
ENST00000366488.4
ENST00000536561.1 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chr15_+_75628419 | 0.34 |
ENST00000567377.1
ENST00000562789.1 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr15_+_69365272 | 0.33 |
ENST00000559914.1
ENST00000558369.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr4_+_37962018 | 0.33 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr21_-_16031122 | 0.33 |
ENST00000400562.1
|
AF165138.7
|
Protein LOC388813 |
| chr15_+_75628394 | 0.32 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr12_+_133657461 | 0.32 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr20_-_18477862 | 0.31 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr8_+_104831440 | 0.31 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr22_-_38239808 | 0.31 |
ENST00000406423.1
ENST00000424350.1 ENST00000458278.2 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr4_-_140223670 | 0.30 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr11_-_64545222 | 0.29 |
ENST00000433274.2
ENST00000432725.1 |
SF1
|
splicing factor 1 |
| chrX_-_47518527 | 0.29 |
ENST00000333119.3
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr17_-_18266797 | 0.29 |
ENST00000316694.3
ENST00000539052.1 |
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr4_+_96012585 | 0.27 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr14_+_39703084 | 0.27 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr12_+_133757995 | 0.27 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr10_+_104614008 | 0.26 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr22_+_29168652 | 0.26 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr19_-_46195029 | 0.26 |
ENST00000588599.1
ENST00000585392.1 ENST00000590212.1 ENST00000587367.1 ENST00000391932.3 |
SNRPD2
|
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa |
| chr7_-_104909435 | 0.26 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr12_-_7656357 | 0.26 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr12_-_25150409 | 0.25 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr7_+_106505912 | 0.25 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr7_-_38313235 | 0.25 |
ENST00000390338.2
|
TRGJP
|
T cell receptor gamma joining P |
| chr18_-_21017817 | 0.24 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr16_-_46649905 | 0.24 |
ENST00000569702.1
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr9_+_131901710 | 0.24 |
ENST00000524946.2
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr5_+_115387163 | 0.24 |
ENST00000601302.2
|
ARL14EPL
|
ADP-ribosylation factor-like 14 effector protein-like |
| chr7_-_92777606 | 0.24 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr11_+_5009424 | 0.24 |
ENST00000300762.1
|
MMP26
|
matrix metallopeptidase 26 |
| chr4_-_83769996 | 0.23 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr3_-_15382875 | 0.23 |
ENST00000408919.3
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr5_+_72509751 | 0.23 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr4_+_95128748 | 0.23 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_+_74171301 | 0.23 |
ENST00000415954.2
ENST00000498286.1 ENST00000370305.1 ENST00000370300.4 |
MTO1
|
mitochondrial tRNA translation optimization 1 |
| chr1_+_12916941 | 0.22 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr8_-_37797621 | 0.22 |
ENST00000524298.1
ENST00000307599.4 |
GOT1L1
|
glutamic-oxaloacetic transaminase 1-like 1 |
| chr10_+_104613980 | 0.22 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr15_-_85197501 | 0.22 |
ENST00000434634.2
|
WDR73
|
WD repeat domain 73 |
| chr6_-_138893661 | 0.22 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chr12_+_58166726 | 0.22 |
ENST00000546504.1
|
RP11-571M6.15
|
Uncharacterized protein |
| chr11_+_120255997 | 0.21 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr9_-_106087316 | 0.21 |
ENST00000411575.1
|
RP11-341A22.2
|
RP11-341A22.2 |
| chr17_-_39341594 | 0.21 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr14_+_22951276 | 0.21 |
ENST00000390484.1
|
TRAJ54
|
T cell receptor alpha joining 54 |
| chr15_-_34880646 | 0.21 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr5_+_68860949 | 0.21 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr10_-_45802907 | 0.20 |
ENST00000374401.2
|
OR13A1
|
olfactory receptor, family 13, subfamily A, member 1 |
| chr12_-_75905374 | 0.20 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr4_+_158493642 | 0.20 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr10_+_134258649 | 0.20 |
ENST00000392630.3
ENST00000321248.2 |
C10orf91
|
chromosome 10 open reading frame 91 |
| chr10_+_118083919 | 0.19 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr12_+_50690489 | 0.19 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr1_+_149239529 | 0.19 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB16
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.0 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 1.2 | 5.9 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.7 | 5.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.7 | 4.9 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.4 | 1.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.4 | 1.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.3 | 1.7 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.3 | 1.4 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.3 | 1.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.3 | 0.8 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.3 | 1.0 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.3 | 2.5 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.2 | 0.9 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.2 | 2.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.7 | GO:0035566 | negative regulation of icosanoid secretion(GO:0032304) regulation of metanephros size(GO:0035566) |
| 0.2 | 0.6 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.2 | 0.7 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 0.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.6 | GO:0050705 | regulation of interleukin-1 alpha secretion(GO:0050705) |
| 0.1 | 0.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 1.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 1.6 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 2.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 1.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 1.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.7 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.1 | 0.4 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 1.4 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.7 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.2 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.1 | 0.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 5.0 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.8 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 1.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.6 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.4 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.6 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 1.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 2.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 1.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.4 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.2 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 1.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 3.0 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.8 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.9 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 11.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.8 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.3 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.0 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.3 | 1.3 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.3 | 5.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 1.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 11.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 7.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 11.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.4 | 1.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.4 | 1.7 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.4 | 2.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.3 | 1.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.3 | 0.8 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.2 | 1.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 1.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.9 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 1.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 1.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 1.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 5.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.6 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 1.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 1.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.5 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.4 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 2.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 1.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 5.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 4.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.0 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 5.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 5.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 8.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 2.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 4.5 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.7 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |