Project
Illumina Body Map 2
Navigation
Downloads
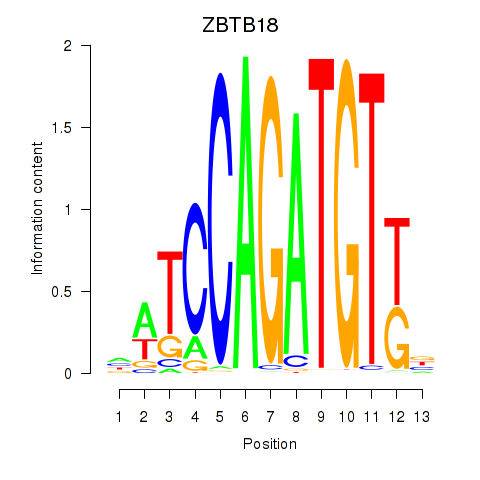
Results for ZBTB18
Z-value: 1.67
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB18
|
ENSG00000179456.9 | zinc finger and BTB domain containing 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB18 | hg19_v2_chr1_+_244214577_244214593 | -0.29 | 1.1e-01 | Click! |
Activity profile of ZBTB18 motif
Sorted Z-values of ZBTB18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_47653178 | 13.13 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr1_-_168698433 | 10.93 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr17_+_47653471 | 10.76 |
ENST00000513748.1
|
NXPH3
|
neurexophilin 3 |
| chr19_+_11649532 | 10.05 |
ENST00000252456.2
ENST00000592923.1 ENST00000535659.2 |
CNN1
|
calponin 1, basic, smooth muscle |
| chr4_+_15376165 | 9.02 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr1_-_16344500 | 7.90 |
ENST00000406363.2
ENST00000411503.1 ENST00000545268.1 ENST00000487046.1 |
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular) |
| chr2_+_189839046 | 7.08 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr11_-_68780824 | 6.95 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chrX_+_99899180 | 5.70 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr17_+_77020325 | 5.53 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77020224 | 5.51 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr15_+_49715449 | 5.48 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr1_+_53527854 | 5.35 |
ENST00000371500.3
ENST00000395871.2 ENST00000312553.5 |
PODN
|
podocan |
| chr2_-_163099885 | 5.26 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr2_-_163100045 | 5.07 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr17_+_77020146 | 5.00 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr14_-_38725573 | 4.81 |
ENST00000342213.2
|
CLEC14A
|
C-type lectin domain family 14, member A |
| chr14_+_94577074 | 4.52 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chrX_+_43515467 | 4.37 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr17_-_15168624 | 4.24 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr13_+_113777105 | 4.21 |
ENST00000409306.1
ENST00000375551.3 ENST00000375559.3 |
F10
|
coagulation factor X |
| chr15_+_49715293 | 4.17 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr16_-_49698136 | 4.08 |
ENST00000535559.1
|
ZNF423
|
zinc finger protein 423 |
| chr15_-_96590126 | 4.01 |
ENST00000561051.1
|
RP11-4G2.1
|
RP11-4G2.1 |
| chr2_-_163099546 | 3.98 |
ENST00000447386.1
|
FAP
|
fibroblast activation protein, alpha |
| chr12_-_104443890 | 3.94 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr11_-_68780701 | 3.88 |
ENST00000320913.6
|
MRGPRF
|
MAS-related GPR, member F |
| chr9_-_13165457 | 3.83 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr12_-_16762802 | 3.78 |
ENST00000534946.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr9_+_72658490 | 3.77 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr12_+_2162447 | 3.71 |
ENST00000335762.5
ENST00000399655.1 |
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chrX_-_101771645 | 3.64 |
ENST00000289373.4
|
TMSB15A
|
thymosin beta 15a |
| chr4_-_152149033 | 3.63 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr1_-_156647189 | 3.58 |
ENST00000368223.3
|
NES
|
nestin |
| chr11_+_76494253 | 3.56 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr3_-_145940214 | 3.42 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr18_+_6834472 | 3.39 |
ENST00000581099.1
ENST00000419673.2 ENST00000531294.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr4_+_75480629 | 3.30 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr11_-_101454658 | 3.30 |
ENST00000344327.3
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr17_-_62050278 | 3.30 |
ENST00000578147.1
ENST00000435607.1 |
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit |
| chr7_+_97361388 | 3.25 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr4_+_75310851 | 3.25 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr17_+_32612687 | 3.17 |
ENST00000305869.3
|
CCL11
|
chemokine (C-C motif) ligand 11 |
| chr19_-_36001286 | 3.07 |
ENST00000602679.1
ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN
|
dermokine |
| chr8_-_49834299 | 3.03 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr4_+_75311019 | 2.99 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr3_-_145940126 | 2.93 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr8_-_17555164 | 2.89 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr1_+_19967014 | 2.88 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr8_+_1772132 | 2.86 |
ENST00000349830.3
ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr12_+_26126681 | 2.76 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr13_-_28545276 | 2.66 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr2_+_177053307 | 2.64 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr7_+_97361218 | 2.63 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr1_+_78354175 | 2.60 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr1_+_78354330 | 2.59 |
ENST00000440324.1
|
NEXN
|
nexilin (F actin binding protein) |
| chr19_-_36001113 | 2.58 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr8_-_49833978 | 2.57 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr20_+_56725952 | 2.50 |
ENST00000371168.3
|
C20orf85
|
chromosome 20 open reading frame 85 |
| chr15_-_74043816 | 2.46 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr2_-_65659762 | 2.45 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr2_+_108994466 | 2.44 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr3_+_100211412 | 2.40 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr17_-_67138015 | 2.27 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr19_-_36001386 | 2.25 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chr1_+_78354297 | 2.25 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr11_-_1606513 | 2.24 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr10_-_105437909 | 2.21 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr8_+_24241969 | 2.18 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr4_-_111563076 | 2.18 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr12_-_48119301 | 2.17 |
ENST00000545824.2
ENST00000422538.3 |
ENDOU
|
endonuclease, polyU-specific |
| chr5_+_140782351 | 2.17 |
ENST00000573521.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chr7_-_135433534 | 2.14 |
ENST00000338588.3
|
FAM180A
|
family with sequence similarity 180, member A |
| chr7_+_100770328 | 2.14 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr6_-_163148700 | 2.13 |
ENST00000366894.1
ENST00000338468.3 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr19_+_41119323 | 2.07 |
ENST00000599724.1
ENST00000597071.1 ENST00000243562.9 |
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr13_+_113699029 | 2.07 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr9_+_87283430 | 2.04 |
ENST00000376214.1
ENST00000376213.1 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr13_-_33780133 | 2.02 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr11_-_119252425 | 2.01 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr8_+_24241789 | 2.01 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr1_+_38022513 | 1.99 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chrX_-_153141302 | 1.93 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chrX_-_154563889 | 1.88 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr11_-_119252359 | 1.86 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr15_+_51669444 | 1.84 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr1_+_38022572 | 1.84 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr4_-_111563279 | 1.83 |
ENST00000511837.1
|
PITX2
|
paired-like homeodomain 2 |
| chr17_+_32582293 | 1.78 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr19_+_54135310 | 1.76 |
ENST00000376650.1
|
DPRX
|
divergent-paired related homeobox |
| chr17_+_48638371 | 1.71 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr9_+_97488939 | 1.71 |
ENST00000277198.2
ENST00000297979.5 |
C9orf3
|
chromosome 9 open reading frame 3 |
| chr1_+_78354243 | 1.70 |
ENST00000294624.8
|
NEXN
|
nexilin (F actin binding protein) |
| chr8_+_38965048 | 1.68 |
ENST00000399831.3
ENST00000437682.2 ENST00000519315.1 ENST00000379907.4 ENST00000522506.1 |
ADAM32
|
ADAM metallopeptidase domain 32 |
| chr17_+_32597232 | 1.68 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr19_+_7584088 | 1.55 |
ENST00000394341.2
|
ZNF358
|
zinc finger protein 358 |
| chr11_-_33722286 | 1.54 |
ENST00000451594.2
ENST00000379011.4 |
C11orf91
|
chromosome 11 open reading frame 91 |
| chr6_-_163148780 | 1.53 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr16_-_18470696 | 1.49 |
ENST00000427999.2
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr12_-_48119340 | 1.48 |
ENST00000229003.3
|
ENDOU
|
endonuclease, polyU-specific |
| chr14_+_38033252 | 1.47 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr3_-_111314230 | 1.42 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr11_+_4673716 | 1.40 |
ENST00000530215.1
|
OR51E1
|
olfactory receptor, family 51, subfamily E, member 1 |
| chrX_+_103031758 | 1.38 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr12_-_16762971 | 1.37 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr17_-_8263538 | 1.36 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr13_+_113698946 | 1.35 |
ENST00000397021.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr11_+_15136462 | 1.35 |
ENST00000379556.3
ENST00000424273.1 |
INSC
|
inscuteable homolog (Drosophila) |
| chr12_+_1738363 | 1.34 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chrX_-_153141434 | 1.33 |
ENST00000407935.2
ENST00000439496.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr17_-_41174424 | 1.33 |
ENST00000355653.3
|
VAT1
|
vesicle amine transport 1 |
| chr17_+_36584662 | 1.32 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr11_+_63870660 | 1.28 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr5_-_180552304 | 1.24 |
ENST00000329365.2
|
OR2V1
|
olfactory receptor, family 2, subfamily V, member 1 |
| chrX_-_110513703 | 1.24 |
ENST00000324068.1
|
CAPN6
|
calpain 6 |
| chr17_-_19651668 | 1.23 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr12_+_77718388 | 1.21 |
ENST00000550042.1
|
RP1-34H18.1
|
RP1-34H18.1 |
| chr7_-_135433460 | 1.17 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr11_-_57089774 | 1.15 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr16_+_28914680 | 1.14 |
ENST00000564112.1
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr17_-_41174367 | 1.13 |
ENST00000587173.1
|
VAT1
|
vesicle amine transport 1 |
| chr3_+_159570722 | 1.13 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr11_+_71900703 | 1.13 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr5_+_140792614 | 1.13 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr1_-_207206092 | 1.12 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chrX_+_103031421 | 1.09 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr2_+_33359687 | 1.09 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_-_19651654 | 1.07 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr11_+_32112431 | 1.07 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr3_-_105588231 | 1.05 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr2_+_90139056 | 1.04 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr12_-_54121261 | 1.03 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr1_+_37947257 | 1.01 |
ENST00000471012.1
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr9_+_118916082 | 1.00 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr2_+_33359646 | 0.98 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_-_102874102 | 0.96 |
ENST00000392905.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr11_-_57089671 | 0.96 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr11_+_5602107 | 0.93 |
ENST00000345043.2
|
OR52B6
|
olfactory receptor, family 52, subfamily B, member 6 |
| chr8_-_11058847 | 0.90 |
ENST00000297303.4
ENST00000416569.2 |
XKR6
|
XK, Kell blood group complex subunit-related family, member 6 |
| chr19_+_38880695 | 0.88 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr12_-_56121612 | 0.88 |
ENST00000546939.1
|
CD63
|
CD63 molecule |
| chr10_-_95241951 | 0.85 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr18_+_3448455 | 0.82 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr12_-_56121580 | 0.78 |
ENST00000550776.1
|
CD63
|
CD63 molecule |
| chr11_-_40315640 | 0.78 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr6_-_39197226 | 0.75 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr3_+_121311966 | 0.75 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr10_-_95242044 | 0.75 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr8_-_37411648 | 0.74 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr12_-_11175219 | 0.72 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr2_-_89399845 | 0.68 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr6_-_30523865 | 0.65 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr1_-_11907829 | 0.64 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr11_+_5841544 | 0.64 |
ENST00000317037.2
|
OR52N2
|
olfactory receptor, family 52, subfamily N, member 2 |
| chr19_+_39903185 | 0.62 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr11_-_63684316 | 0.61 |
ENST00000301459.4
|
RCOR2
|
REST corepressor 2 |
| chr7_-_156803329 | 0.60 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr3_-_114035026 | 0.54 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr2_+_29341037 | 0.54 |
ENST00000449202.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr19_+_50084561 | 0.53 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr1_-_32210275 | 0.53 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr14_-_61748550 | 0.50 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr15_+_51669513 | 0.50 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chr1_-_113160826 | 0.50 |
ENST00000538187.1
ENST00000369664.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr15_-_43559055 | 0.50 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr10_+_97733786 | 0.49 |
ENST00000371198.2
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr19_-_13947099 | 0.47 |
ENST00000587762.1
|
MIR24-2
|
microRNA 24-2 |
| chr11_-_5810046 | 0.46 |
ENST00000317078.1
|
OR52N1
|
olfactory receptor, family 52, subfamily N, member 1 |
| chr4_+_144312659 | 0.45 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr14_+_67707826 | 0.44 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr2_+_33359473 | 0.43 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr15_+_47631257 | 0.42 |
ENST00000560636.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr6_+_163148973 | 0.40 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr10_-_123357598 | 0.38 |
ENST00000358487.5
ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr12_-_56122220 | 0.38 |
ENST00000552692.1
|
CD63
|
CD63 molecule |
| chr17_-_19651598 | 0.37 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr20_+_57875658 | 0.36 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr10_-_71332994 | 0.36 |
ENST00000242462.4
|
NEUROG3
|
neurogenin 3 |
| chr3_-_99245957 | 0.35 |
ENST00000484675.1
|
RP11-779P15.2
|
RP11-779P15.2 |
| chr9_+_131683174 | 0.35 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr15_-_74501310 | 0.32 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr12_-_56122124 | 0.30 |
ENST00000552754.1
|
CD63
|
CD63 molecule |
| chr11_+_5905501 | 0.28 |
ENST00000316987.2
|
OR52E4
|
olfactory receptor, family 52, subfamily E, member 4 |
| chr17_+_19282064 | 0.26 |
ENST00000603493.1
|
MAPK7
|
mitogen-activated protein kinase 7 |
| chr2_-_89417335 | 0.26 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr2_+_201994042 | 0.26 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr15_+_67418047 | 0.25 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr19_+_48673949 | 0.25 |
ENST00000328759.7
|
C19orf68
|
chromosome 19 open reading frame 68 |
| chr2_-_89597542 | 0.24 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr2_+_89923550 | 0.22 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr15_-_31453359 | 0.21 |
ENST00000542188.1
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr11_+_5509915 | 0.19 |
ENST00000322641.5
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1 |
| chr17_+_64961026 | 0.18 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr16_+_14564842 | 0.17 |
ENST00000599734.1
|
AC092291.2
|
AC092291.2 |
| chr20_+_30467600 | 0.17 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr19_-_15918936 | 0.17 |
ENST00000334920.2
|
OR10H1
|
olfactory receptor, family 10, subfamily H, member 1 |
| chr8_+_38662960 | 0.16 |
ENST00000524193.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr1_+_42619070 | 0.16 |
ENST00000372581.1
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin) |
| chr1_+_248651885 | 0.16 |
ENST00000366473.2
|
OR2T5
|
olfactory receptor, family 2, subfamily T, member 5 |
| chr19_+_9203855 | 0.16 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 3.2 | 9.6 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 2.0 | 16.0 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 2.0 | 5.9 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 1.9 | 5.6 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.1 | 7.9 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.9 | 3.6 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.8 | 3.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.8 | 4.0 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.8 | 7.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.7 | 2.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.6 | 0.6 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.6 | 3.2 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.6 | 6.2 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.6 | 1.8 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.6 | 2.9 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.6 | 2.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.5 | 4.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 3.7 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.4 | 10.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.4 | 1.1 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.4 | 1.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.4 | 5.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.4 | 2.5 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.4 | 9.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 1.7 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.3 | 2.0 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.3 | 2.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 1.0 | GO:2000625 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.3 | 5.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 2.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 4.4 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.3 | 2.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 6.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 1.7 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 2.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 4.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 10.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 3.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.0 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.2 | 2.5 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 23.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.2 | 3.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.1 | 0.4 | GO:0060615 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.1 | 5.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 3.6 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.1 | 2.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 2.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.9 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 1.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.3 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.1 | 2.9 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 2.4 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.1 | 4.5 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 5.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 4.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 3.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.3 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 1.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 1.0 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 1.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.0 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 3.9 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 1.5 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.4 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.0 | 8.3 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 2.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 2.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0014826 | cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 2.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 3.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.5 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 1.4 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 2.0 | GO:0030449 | regulation of complement activation(GO:0030449) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 14.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.5 | 7.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 3.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 8.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 3.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 27.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 5.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 2.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 3.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.0 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 2.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 7.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.3 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 6.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 3.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 4.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 22.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 10.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 8.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 6.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 6.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 6.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 5.8 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 2.1 | GO:0043231 | intracellular membrane-bounded organelle(GO:0043231) |
| 0.0 | 2.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 1.6 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 3.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.6 | 2.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 5.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.5 | 4.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 2.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 14.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.4 | 1.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 2.0 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.3 | 7.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 10.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 2.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 1.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.3 | 6.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 7.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 2.9 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 3.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 2.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 3.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 3.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 2.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 21.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 6.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 4.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 3.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 3.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 6.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 2.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 8.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 3.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 4.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 6.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 2.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 10.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 4.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 6.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.5 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 3.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 2.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 3.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 28.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 7.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 4.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 8.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 25.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.7 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 3.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 3.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.8 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 3.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 6.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 6.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 4.8 | NABA MATRISOME | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.0 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 4.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 4.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 6.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 8.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 6.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 3.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 4.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 3.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 6.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 3.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |