Project
Illumina Body Map 2
Navigation
Downloads
Results for ZBTB49
Z-value: 0.66
Transcription factors associated with ZBTB49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB49
|
ENSG00000168826.11 | zinc finger and BTB domain containing 49 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB49 | hg19_v2_chr4_+_4291924_4292011 | -0.03 | 8.7e-01 | Click! |
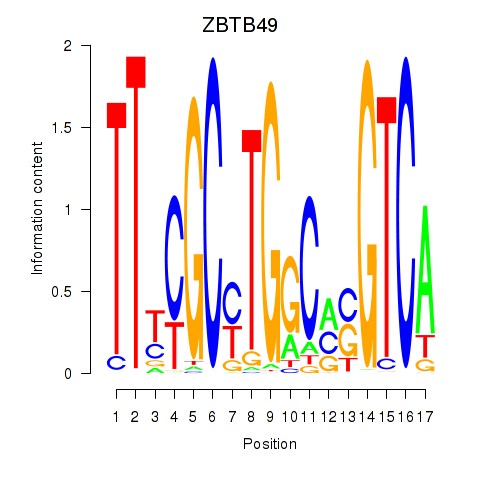
Activity profile of ZBTB49 motif
Sorted Z-values of ZBTB49 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_95691445 | 1.68 |
ENST00000353004.3
ENST00000354078.3 ENST00000349807.3 |
MAL
|
mal, T-cell differentiation protein |
| chr15_+_90728145 | 1.60 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr2_+_95691417 | 1.46 |
ENST00000309988.4
|
MAL
|
mal, T-cell differentiation protein |
| chr19_-_6481776 | 1.40 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr8_-_82024290 | 1.29 |
ENST00000220597.4
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr19_-_6481759 | 1.25 |
ENST00000588421.1
|
DENND1C
|
DENN/MADD domain containing 1C |
| chr12_+_40618764 | 1.24 |
ENST00000343742.2
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr1_+_26869597 | 1.23 |
ENST00000530003.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr12_+_40618873 | 1.19 |
ENST00000298910.7
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr11_+_117857063 | 1.18 |
ENST00000227752.3
ENST00000541785.1 ENST00000545409.1 |
IL10RA
|
interleukin 10 receptor, alpha |
| chr15_+_31508174 | 1.11 |
ENST00000559292.2
ENST00000557928.1 |
RP11-16E12.1
|
RP11-16E12.1 |
| chr9_-_97401782 | 1.06 |
ENST00000375326.4
|
FBP1
|
fructose-1,6-bisphosphatase 1 |
| chr16_-_28621353 | 0.92 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr18_+_32558380 | 0.81 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr3_-_172428842 | 0.78 |
ENST00000424772.1
|
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr17_+_54671047 | 0.77 |
ENST00000332822.4
|
NOG
|
noggin |
| chrX_-_153210107 | 0.77 |
ENST00000369997.3
ENST00000393700.3 ENST00000412763.1 |
RENBP
|
renin binding protein |
| chr2_+_30670209 | 0.74 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr3_-_172428959 | 0.72 |
ENST00000475381.1
ENST00000538775.1 ENST00000273512.3 ENST00000543711.1 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr2_+_89890533 | 0.71 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr16_+_10972818 | 0.68 |
ENST00000576601.1
|
CIITA
|
class II, major histocompatibility complex, transactivator |
| chr18_+_32558208 | 0.66 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr6_-_6320875 | 0.63 |
ENST00000451619.1
|
F13A1
|
coagulation factor XIII, A1 polypeptide |
| chr11_+_108094786 | 0.63 |
ENST00000601453.1
|
ATM
|
ataxia telangiectasia mutated |
| chr3_-_196242233 | 0.62 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr9_-_123676827 | 0.62 |
ENST00000546084.1
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr4_+_128886424 | 0.60 |
ENST00000398965.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr12_+_83080724 | 0.59 |
ENST00000548305.1
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr6_-_159240415 | 0.57 |
ENST00000367075.3
|
EZR
|
ezrin |
| chr3_+_196669494 | 0.55 |
ENST00000602845.1
|
NCBP2-AS2
|
NCBP2 antisense RNA 2 (head to head) |
| chrX_-_131623874 | 0.53 |
ENST00000436215.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr14_+_35591509 | 0.52 |
ENST00000604073.1
|
KIAA0391
|
KIAA0391 |
| chr18_+_43753500 | 0.51 |
ENST00000587591.1
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr14_-_93673353 | 0.50 |
ENST00000556566.1
ENST00000306954.4 |
C14orf142
|
chromosome 14 open reading frame 142 |
| chr2_-_61244550 | 0.48 |
ENST00000421319.1
|
PUS10
|
pseudouridylate synthase 10 |
| chr8_+_24151553 | 0.46 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr2_+_30670127 | 0.45 |
ENST00000540623.1
ENST00000476038.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr3_+_52232102 | 0.44 |
ENST00000469224.1
ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr22_-_29196511 | 0.43 |
ENST00000344347.5
|
XBP1
|
X-box binding protein 1 |
| chr3_-_184971817 | 0.43 |
ENST00000440662.1
ENST00000456310.1 |
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr14_+_35591858 | 0.42 |
ENST00000603544.1
|
KIAA0391
|
KIAA0391 |
| chr1_+_79115503 | 0.41 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr2_-_179659669 | 0.40 |
ENST00000436599.1
|
TTN
|
titin |
| chrX_+_64887512 | 0.39 |
ENST00000360270.5
|
MSN
|
moesin |
| chr12_+_50505762 | 0.39 |
ENST00000550487.1
ENST00000317943.2 |
COX14
|
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
| chr10_+_90750493 | 0.38 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr14_+_35591928 | 0.38 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr1_-_151319283 | 0.37 |
ENST00000392746.3
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr12_+_83080659 | 0.37 |
ENST00000321196.3
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr5_+_154238096 | 0.36 |
ENST00000517568.1
ENST00000524105.1 ENST00000285896.6 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr8_+_99129513 | 0.36 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chrX_+_9431324 | 0.35 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr2_+_30670077 | 0.35 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr20_-_25320367 | 0.35 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chrX_+_101854426 | 0.33 |
ENST00000536530.1
ENST00000473968.1 ENST00000604957.1 ENST00000477663.1 ENST00000479502.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr6_-_49604545 | 0.33 |
ENST00000371175.4
ENST00000229810.7 |
RHAG
|
Rh-associated glycoprotein |
| chr12_+_53835539 | 0.32 |
ENST00000547368.1
ENST00000379786.4 ENST00000551945.1 |
PRR13
|
proline rich 13 |
| chrX_+_101854096 | 0.32 |
ENST00000246174.2
ENST00000537008.1 ENST00000541409.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr22_-_29196030 | 0.31 |
ENST00000405219.3
|
XBP1
|
X-box binding protein 1 |
| chr2_-_27498208 | 0.31 |
ENST00000424577.1
ENST00000426569.1 |
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr10_+_81466084 | 0.31 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr4_-_53525406 | 0.29 |
ENST00000451218.2
ENST00000441222.3 |
USP46
|
ubiquitin specific peptidase 46 |
| chr12_+_53835425 | 0.28 |
ENST00000549924.1
|
PRR13
|
proline rich 13 |
| chr10_-_46641003 | 0.28 |
ENST00000395721.2
ENST00000374218.2 ENST00000395725.3 ENST00000374346.3 ENST00000417004.1 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr14_+_35591735 | 0.28 |
ENST00000604948.1
ENST00000605201.1 ENST00000250377.7 ENST00000321130.10 ENST00000534898.4 |
KIAA0391
|
KIAA0391 |
| chr5_+_154237778 | 0.28 |
ENST00000523698.1
ENST00000517876.1 ENST00000520472.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr5_+_154238149 | 0.27 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr15_-_25684110 | 0.27 |
ENST00000232165.3
|
UBE3A
|
ubiquitin protein ligase E3A |
| chr16_+_2533020 | 0.26 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr8_-_42396632 | 0.26 |
ENST00000520179.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr2_-_27498186 | 0.25 |
ENST00000447008.2
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr22_-_20255212 | 0.25 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr16_+_30406423 | 0.24 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr2_+_95873257 | 0.24 |
ENST00000425953.1
|
AC092835.2
|
AC092835.2 |
| chr12_+_53835508 | 0.24 |
ENST00000551003.1
ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13
PCBP2
|
proline rich 13 poly(rC) binding protein 2 |
| chr8_-_99129338 | 0.24 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr7_+_99006232 | 0.23 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr2_-_85555086 | 0.23 |
ENST00000444342.2
ENST00000409232.3 ENST00000409015.1 |
TGOLN2
|
trans-golgi network protein 2 |
| chr22_-_29196546 | 0.23 |
ENST00000403532.3
ENST00000216037.6 |
XBP1
|
X-box binding protein 1 |
| chr11_+_63993738 | 0.22 |
ENST00000441250.2
ENST00000279206.3 |
NUDT22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr5_+_154238042 | 0.22 |
ENST00000519211.1
ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr19_-_54567035 | 0.22 |
ENST00000366170.2
ENST00000425006.2 |
VSTM1
|
V-set and transmembrane domain containing 1 |
| chr12_+_53835383 | 0.22 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr8_-_99129384 | 0.21 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr7_+_148936732 | 0.21 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr19_+_17862274 | 0.21 |
ENST00000596536.1
ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1
|
FCH domain only 1 |
| chr12_-_120241187 | 0.21 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr1_-_151319318 | 0.21 |
ENST00000436271.1
ENST00000450506.1 ENST00000422595.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr14_-_74485960 | 0.21 |
ENST00000556242.1
ENST00000334696.6 |
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr16_+_30406721 | 0.19 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr20_-_33735070 | 0.19 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr22_-_42017021 | 0.19 |
ENST00000263256.6
|
DESI1
|
desumoylating isopeptidase 1 |
| chr19_+_29097595 | 0.18 |
ENST00000587961.1
|
AC079466.1
|
AC079466.1 |
| chr2_-_85555355 | 0.17 |
ENST00000282120.2
ENST00000398263.2 |
TGOLN2
|
trans-golgi network protein 2 |
| chr16_-_18937480 | 0.17 |
ENST00000532700.2
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr22_-_20256054 | 0.17 |
ENST00000043402.7
|
RTN4R
|
reticulon 4 receptor |
| chr5_-_77590480 | 0.17 |
ENST00000519295.1
ENST00000255194.6 |
AP3B1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr7_-_44613494 | 0.17 |
ENST00000431640.1
ENST00000258772.5 |
DDX56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr4_+_144312659 | 0.16 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr12_+_104324112 | 0.16 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr1_+_182808474 | 0.15 |
ENST00000367549.3
|
DHX9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr1_+_24117627 | 0.15 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr5_-_180229833 | 0.14 |
ENST00000307826.4
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr16_-_18937726 | 0.14 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_-_151319654 | 0.12 |
ENST00000430227.1
ENST00000412774.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr19_-_4182530 | 0.12 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr19_-_51611623 | 0.11 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr10_+_64893039 | 0.09 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr14_-_23446900 | 0.09 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr21_-_26979786 | 0.08 |
ENST00000419219.1
ENST00000352957.4 ENST00000307301.7 |
MRPL39
|
mitochondrial ribosomal protein L39 |
| chr20_+_5731083 | 0.08 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr22_+_21996549 | 0.06 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr14_-_35591679 | 0.06 |
ENST00000557278.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr7_-_12443501 | 0.06 |
ENST00000275358.3
|
VWDE
|
von Willebrand factor D and EGF domains |
| chr1_+_8021954 | 0.06 |
ENST00000377491.1
ENST00000377488.1 |
PARK7
|
parkinson protein 7 |
| chr3_+_196594727 | 0.06 |
ENST00000445299.2
ENST00000323460.5 ENST00000419026.1 |
SENP5
|
SUMO1/sentrin specific peptidase 5 |
| chr19_-_51466681 | 0.06 |
ENST00000456750.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr17_+_74734052 | 0.06 |
ENST00000590514.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr11_+_43702322 | 0.05 |
ENST00000395700.4
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr14_-_35591433 | 0.05 |
ENST00000261475.5
ENST00000555644.1 |
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr8_-_59572093 | 0.04 |
ENST00000427130.2
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr11_-_63993690 | 0.04 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr14_-_35591156 | 0.04 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr4_+_53525573 | 0.02 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr6_-_109776901 | 0.01 |
ENST00000431946.1
|
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr1_-_36863481 | 0.00 |
ENST00000315732.2
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB49
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:1903217 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.3 | 1.0 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) epithelial cell maturation involved in salivary gland development(GO:0060691) regulation of plasma cell differentiation(GO:1900098) positive regulation of plasma cell differentiation(GO:1900100) positive regulation of lactation(GO:1903489) |
| 0.3 | 1.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.2 | 3.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.2 | 1.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.2 | 0.8 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 | 0.8 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 1.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.6 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 1.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 1.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.3 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.1 | 0.3 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.7 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.1 | 0.1 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.4 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.4 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.2 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 1.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 1.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.5 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.2 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0044753 | amphisome(GO:0044753) |
| 0.2 | 1.6 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.6 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 1.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.3 | 1.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 1.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 0.6 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.2 | 3.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.4 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 2.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.4 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.2 | GO:0033677 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) triplex DNA binding(GO:0045142) |
| 0.0 | 1.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |