Project
Illumina Body Map 2
Navigation
Downloads
Results for ZFHX3
Z-value: 1.03
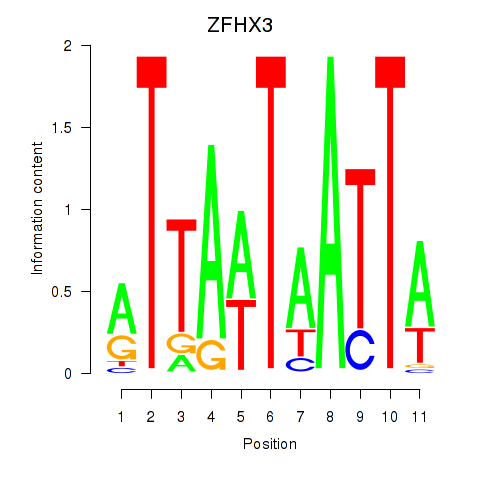
Transcription factors associated with ZFHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFHX3
|
ENSG00000140836.10 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFHX3 | hg19_v2_chr16_-_73082274_73082274 | -0.69 | 1.3e-05 | Click! |
Activity profile of ZFHX3 motif
Sorted Z-values of ZFHX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_63063152 | 9.58 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr4_-_155511887 | 9.00 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr17_-_64225508 | 7.13 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr1_+_159557607 | 6.66 |
ENST00000255040.2
|
APCS
|
amyloid P component, serum |
| chr2_+_234621551 | 5.61 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr14_+_95027772 | 5.53 |
ENST00000555095.1
ENST00000298841.5 ENST00000554220.1 ENST00000553780.1 |
SERPINA4
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr17_-_26697304 | 5.24 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr1_-_159684371 | 5.11 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr17_+_41052808 | 5.07 |
ENST00000592383.1
ENST00000253801.2 ENST00000585489.1 |
G6PC
|
glucose-6-phosphatase, catalytic subunit |
| chr6_+_161123270 | 4.75 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr2_+_234637754 | 4.18 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_+_234627424 | 4.18 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr6_+_50061315 | 4.10 |
ENST00000415106.1
|
RP11-397G17.1
|
RP11-397G17.1 |
| chr4_+_74347400 | 3.91 |
ENST00000226355.3
|
AFM
|
afamin |
| chr19_-_36304201 | 3.86 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr11_+_22696314 | 3.58 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr9_-_123812542 | 3.36 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr20_-_7921090 | 3.29 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr19_-_58864848 | 3.25 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr15_+_58724184 | 3.19 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr4_+_100495864 | 3.09 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr17_+_4692230 | 3.01 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr2_+_88047606 | 2.81 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr6_+_25754927 | 2.73 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr7_-_44580861 | 2.43 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr6_-_161085291 | 2.34 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr1_-_197036364 | 2.04 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr2_-_87248975 | 2.04 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr4_+_3443614 | 1.98 |
ENST00000382774.3
ENST00000511533.1 |
HGFAC
|
HGF activator |
| chr3_+_46919235 | 1.90 |
ENST00000449590.1
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr20_+_42187682 | 1.81 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr11_-_111649074 | 1.72 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr20_+_42187608 | 1.69 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chrX_-_32173579 | 1.65 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr7_-_7575477 | 1.49 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chrM_+_8527 | 1.49 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chrX_-_15288154 | 1.40 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr1_-_150944411 | 1.28 |
ENST00000368949.4
|
CERS2
|
ceramide synthase 2 |
| chrM_-_14670 | 1.27 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr14_-_104408645 | 1.23 |
ENST00000557640.1
|
RD3L
|
retinal degeneration 3-like |
| chrM_+_10464 | 1.20 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr20_-_33735070 | 1.19 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr5_-_125930929 | 1.14 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chrM_+_9207 | 1.11 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr4_-_22444733 | 1.09 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr5_-_115890554 | 1.08 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr9_-_95298314 | 1.04 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr16_-_69385681 | 1.01 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr15_-_76352069 | 0.99 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr1_-_153585539 | 0.94 |
ENST00000368706.4
|
S100A16
|
S100 calcium binding protein A16 |
| chrM_+_14741 | 0.94 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr8_-_145754428 | 0.93 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr12_-_7818474 | 0.92 |
ENST00000229304.4
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr7_+_150725510 | 0.83 |
ENST00000461373.1
ENST00000358849.4 ENST00000297504.6 ENST00000542328.1 ENST00000498578.1 ENST00000356058.4 ENST00000477719.1 ENST00000477092.1 |
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr4_-_152149033 | 0.81 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr12_+_59989791 | 0.80 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr5_+_95066823 | 0.77 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr12_+_59989918 | 0.77 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr8_-_18541603 | 0.73 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chrX_-_110655306 | 0.64 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr11_-_58378759 | 0.62 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chr9_-_95298254 | 0.56 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr11_-_559377 | 0.53 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr3_+_77088989 | 0.52 |
ENST00000461745.1
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chrX_+_84258832 | 0.50 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr11_-_61687739 | 0.49 |
ENST00000531922.1
ENST00000301773.5 |
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr12_-_131200810 | 0.46 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chrM_+_10053 | 0.43 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr1_+_212475148 | 0.38 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr10_-_74283694 | 0.38 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr12_+_80750134 | 0.35 |
ENST00000546620.1
|
OTOGL
|
otogelin-like |
| chr1_-_227505289 | 0.35 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr7_+_130126165 | 0.33 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr7_+_130126012 | 0.33 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr4_-_69111401 | 0.30 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr5_+_60933634 | 0.29 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr14_+_39703112 | 0.29 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr17_-_39156138 | 0.27 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr2_+_105050794 | 0.26 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr6_-_31681839 | 0.25 |
ENST00000409239.1
ENST00000461287.1 |
LY6G6E
XXbac-BPG32J3.20
|
lymphocyte antigen 6 complex, locus G6E (pseudogene) Uncharacterized protein |
| chr20_-_21494654 | 0.25 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr3_-_114477787 | 0.21 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr14_-_83262540 | 0.19 |
ENST00000554451.1
|
RP11-11K13.1
|
RP11-11K13.1 |
| chr8_+_104831440 | 0.17 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_192544857 | 0.17 |
ENST00000367459.3
ENST00000469578.2 |
RGS1
|
regulator of G-protein signaling 1 |
| chr22_-_30642728 | 0.15 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr11_+_71934962 | 0.14 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr11_+_5410607 | 0.14 |
ENST00000328611.3
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1 |
| chr7_-_71868354 | 0.13 |
ENST00000412588.1
|
CALN1
|
calneuron 1 |
| chr11_+_57559005 | 0.13 |
ENST00000534647.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr4_-_149365827 | 0.12 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr2_-_40680578 | 0.11 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr11_-_111649015 | 0.10 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr12_+_53835539 | 0.09 |
ENST00000547368.1
ENST00000379786.4 ENST00000551945.1 |
PRR13
|
proline rich 13 |
| chr1_-_47017199 | 0.08 |
ENST00000481882.2
|
KNCN
|
kinocilin |
| chr10_+_118083919 | 0.06 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr1_-_158369256 | 0.06 |
ENST00000334438.1
|
OR10T2
|
olfactory receptor, family 10, subfamily T, member 2 |
| chr12_-_101801505 | 0.05 |
ENST00000539055.1
ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1
|
ADP-ribosylation factor-like 1 |
| chr8_-_141774467 | 0.04 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr18_+_20494078 | 0.04 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr8_-_9762871 | 0.01 |
ENST00000521242.1
|
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chr5_-_35195338 | 0.01 |
ENST00000509839.1
|
PRLR
|
prolactin receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFHX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 14.0 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 1.2 | 4.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.0 | 9.6 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.0 | 6.7 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.9 | 5.5 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.6 | 9.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.6 | 10.2 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.6 | 3.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.6 | 3.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 5.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 3.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.3 | 1.3 | GO:0014011 | Schwann cell proliferation involved in axon regeneration(GO:0014011) negative regulation of Schwann cell migration(GO:1900148) regulation of Schwann cell proliferation involved in axon regeneration(GO:1905044) negative regulation of Schwann cell proliferation involved in axon regeneration(GO:1905045) |
| 0.3 | 0.9 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.3 | 1.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.3 | 5.5 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.3 | 5.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 5.1 | GO:0008228 | opsonization(GO:0008228) positive regulation of superoxide anion generation(GO:0032930) |
| 0.2 | 1.2 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.2 | 1.6 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 2.4 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 1.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 2.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.9 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 3.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.4 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.4 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 1.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 1.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 2.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 3.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 3.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 3.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0036024 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 1.3 | 5.2 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.6 | 9.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 4.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 7.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.4 | 9.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 3.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 5.5 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.1 | 1.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 13.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 3.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 4.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 5.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 9.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 1.3 | 5.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.2 | 11.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.1 | 3.3 | GO:0052853 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 1.0 | 7.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.0 | 3.9 | GO:0008431 | vitamin E binding(GO:0008431) |
| 1.0 | 3.9 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.6 | 5.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 1.6 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.4 | 1.9 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 1.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 9.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.3 | 14.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 5.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 5.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.9 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 3.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 2.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 3.1 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 8.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 1.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 18.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 5.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 9.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 3.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 5.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.3 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 2.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 14.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 5.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 6.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 4.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.3 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 3.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 6.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 5.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 4.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 4.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |