Project
Illumina Body Map 2
Navigation
Downloads
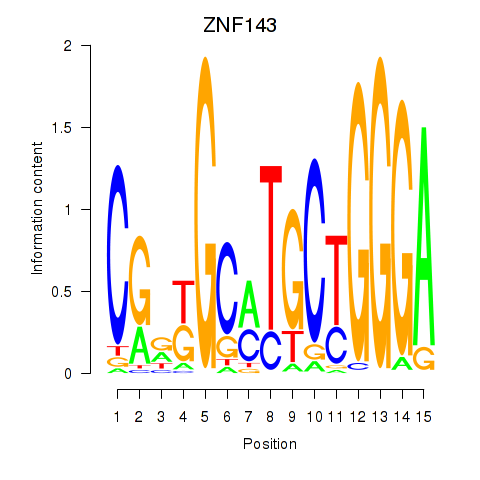
Results for ZNF143
Z-value: 1.90
Transcription factors associated with ZNF143
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF143
|
ENSG00000166478.5 | zinc finger protein 143 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF143 | hg19_v2_chr11_+_9482551_9482604 | -0.10 | 5.8e-01 | Click! |
Activity profile of ZNF143 motif
Sorted Z-values of ZNF143 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF143
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.3 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 1.5 | 4.6 | GO:0060752 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 1.0 | 3.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 1.0 | 2.9 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.8 | 2.5 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.8 | 2.5 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.7 | 2.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.6 | 1.9 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.6 | 2.4 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.6 | 2.9 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.6 | 2.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.6 | 1.7 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.6 | 1.7 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.5 | 3.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 1.1 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.5 | 5.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.5 | 2.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.5 | 3.0 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.5 | 2.0 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.5 | 3.3 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.5 | 1.4 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 | 1.8 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.4 | 1.7 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.7 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.4 | 2.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.4 | 4.8 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.4 | 6.0 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.4 | 1.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.4 | 2.0 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.3 | 1.0 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 1.0 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.3 | 1.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 2.9 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.3 | 4.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 3.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.3 | 0.9 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.3 | 0.9 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.3 | 2.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.3 | 2.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.3 | 1.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.3 | 1.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 0.9 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.3 | 2.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.3 | 1.6 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.3 | 3.5 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.3 | 1.3 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 0.3 | 1.8 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.3 | 0.8 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.2 | 3.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 2.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 2.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.5 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.2 | 1.6 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.2 | 4.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 0.8 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 2.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.6 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.2 | 0.6 | GO:0030474 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.2 | 1.2 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.8 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 1.2 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 2.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 2.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.2 | 2.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 2.9 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 1.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 0.9 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 1.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 2.8 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.2 | 1.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 1.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 1.7 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 0.5 | GO:0052552 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.2 | 0.5 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.2 | 0.8 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 1.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 3.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.2 | 0.6 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.2 | 0.5 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 1.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.2 | 0.9 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.9 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 2.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 1.9 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 1.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 1.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.5 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.1 | 3.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.4 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.7 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.8 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 3.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 1.0 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 2.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 1.8 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.9 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 2.2 | GO:0039529 | RIG-I signaling pathway(GO:0039529) |
| 0.1 | 2.1 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.1 | 0.7 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 1.4 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 | 3.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.6 | GO:0009304 | tRNA transcription(GO:0009304) 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 6.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.4 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 1.1 | GO:0036289 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) peptidyl-serine autophosphorylation(GO:0036289) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) insulin metabolic process(GO:1901142) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 2.4 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 1.0 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.0 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.6 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.7 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 3.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.9 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 1.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.5 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 3.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 1.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.4 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.5 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 2.6 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.1 | 0.8 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 1.1 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 | 1.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.2 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.7 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.7 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.4 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 1.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.7 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.1 | 0.8 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 1.0 | GO:0030638 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.1 | 2.6 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.1 | 0.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.9 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.8 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.6 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.4 | GO:0035519 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.5 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 1.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 1.9 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.6 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.1 | 0.8 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 1.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.9 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 1.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.2 | GO:0006914 | autophagy(GO:0006914) |
| 0.1 | 0.1 | GO:1904351 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.1 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 1.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.0 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.5 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.3 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 1.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.3 | GO:1990569 | GDP-fucose transport(GO:0015783) UDP-N-acetylglucosamine transport(GO:0015788) purine nucleotide-sugar transport(GO:0036079) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.2 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.7 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.9 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.3 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.8 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 5.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.7 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.3 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 2.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 2.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.8 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 1.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0030522 | intracellular receptor signaling pathway(GO:0030522) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.7 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.0 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 3.3 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 3.0 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0090340 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.0 | 0.8 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 2.2 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.6 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 1.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.7 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.6 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 1.0 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 1.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 1.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.5 | GO:1900016 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.5 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.7 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 0.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.4 | GO:1904869 | regulation of establishment of protein localization to telomere(GO:0070203) protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 1.5 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.4 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 1.2 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 2.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 2.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 2.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.2 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.8 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 2.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 1.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 1.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 3.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0035196 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.6 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 0.9 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.8 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.2 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.2 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0015781 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 1.3 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.2 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.5 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:2000142 | regulation of DNA-templated transcription, initiation(GO:2000142) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.7 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.4 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.7 | GO:0008038 | neuron recognition(GO:0008038) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0030689 | Noc complex(GO:0030689) |
| 0.7 | 2.9 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.6 | 3.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.6 | 4.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.6 | 2.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.5 | 5.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 2.0 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.5 | 6.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.4 | 6.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.4 | 2.9 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.4 | 2.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 1.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 1.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 1.0 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.3 | 1.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 3.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 3.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 1.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.3 | 4.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 1.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 2.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 0.7 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.2 | 3.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 0.9 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 0.6 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.2 | 1.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 0.6 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 0.8 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 0.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 2.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 2.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 1.6 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 0.5 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.1 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.7 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 1.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 3.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.9 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.7 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.6 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 0.3 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.1 | 1.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 4.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 1.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 3.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.6 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.4 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.6 | GO:0043601 | nuclear replisome(GO:0043601) |
| 0.1 | 1.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.7 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 2.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 5.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.3 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 2.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.4 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 1.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 4.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 1.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 2.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0030894 | replisome(GO:0030894) |
| 0.0 | 5.5 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 2.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 5.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 2.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) pseudopodium(GO:0031143) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 4.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.8 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.1 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 2.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 4.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 1.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 3.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 3.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 3.9 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 2.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 5.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.1 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 1.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 1.0 | 2.9 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.8 | 3.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.7 | 3.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.7 | 2.8 | GO:0004644 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.6 | 1.9 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.6 | 1.9 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.6 | 4.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.6 | 6.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.6 | 1.7 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.6 | 1.7 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.6 | 1.7 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.6 | 1.7 | GO:0042356 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.5 | 3.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.5 | 5.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.5 | 1.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.4 | 1.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.4 | 1.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.4 | 3.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.4 | 1.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.4 | 1.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.3 | 1.0 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.3 | 1.2 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.3 | 2.4 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.3 | 0.9 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 1.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 1.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.3 | 1.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.3 | 1.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.3 | 1.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.3 | 1.6 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.3 | 2.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 1.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 2.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 1.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.2 | 0.9 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 0.7 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.2 | 0.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 0.8 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.2 | 2.8 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.2 | 0.8 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.2 | 1.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 1.0 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.2 | 2.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 2.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 0.9 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 3.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 2.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 1.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 0.5 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.2 | 0.7 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 5.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.6 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 4.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.5 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 2.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 1.9 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.8 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 0.6 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 1.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.5 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.4 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 1.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.5 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.7 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.2 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.1 | 0.4 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 3.8 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.1 | 0.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 1.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.8 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 3.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 2.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.4 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 0.8 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 1.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.3 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 0.3 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 1.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 3.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 1.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 2.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0051998 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 1.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.2 | GO:0034039 | 8-oxo-7,8-dihydroguanine DNA N-glycosylase activity(GO:0034039) |
| 0.1 | 0.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 2.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.6 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.2 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 2.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.3 | GO:0005462 | GDP-fucose transmembrane transporter activity(GO:0005457) UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.2 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 1.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 3.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.8 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 2.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.0 | 0.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 4.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0000701 | purine-specific mismatch base pair DNA N-glycosylase activity(GO:0000701) |
| 0.0 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 2.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 2.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 4.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 5.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 3.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 2.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 1.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 4.9 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 1.0 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.0 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.8 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 5.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.4 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.7 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 1.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 3.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 6.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 3.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 7.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 4.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 4.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.7 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 6.2 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 0.7 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 0.9 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 2.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 3.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 4.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.2 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 5.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.0 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 6.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 2.3 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 4.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 4.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 2.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 4.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 3.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 3.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 3.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.1 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 1.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME HIV LIFE CYCLE | Genes involved in HIV Life Cycle |
| 0.0 | 0.5 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 2.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 3.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |