Project
Illumina Body Map 2
Navigation
Downloads
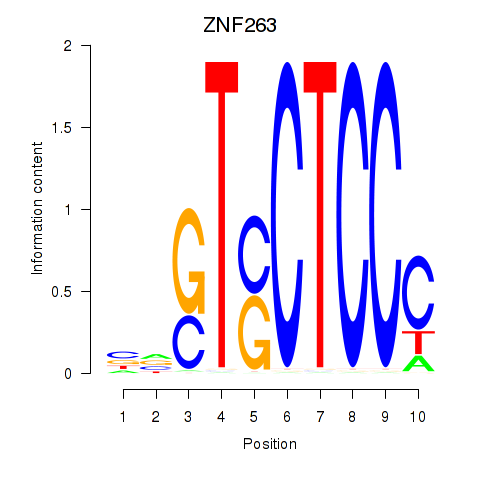
Results for ZNF263
Z-value: 1.43
Transcription factors associated with ZNF263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF263
|
ENSG00000006194.6 | zinc finger protein 263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF263 | hg19_v2_chr16_+_3333443_3333545 | -0.08 | 6.5e-01 | Click! |
Activity profile of ZNF263 motif
Sorted Z-values of ZNF263 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_27066636 | 4.58 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr7_-_150038704 | 3.32 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr19_-_47975417 | 2.94 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr15_-_71146480 | 2.83 |
ENST00000299213.8
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr4_-_154710210 | 2.78 |
ENST00000274063.4
|
SFRP2
|
secreted frizzled-related protein 2 |
| chr19_-_47975106 | 2.69 |
ENST00000539381.1
ENST00000594353.1 ENST00000542837.1 |
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr14_-_27066960 | 2.57 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr5_-_146833803 | 2.51 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr2_-_50574856 | 2.49 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr5_-_146833485 | 2.48 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr19_-_55658687 | 2.48 |
ENST00000593046.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr5_-_146833222 | 2.45 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chrX_-_73072534 | 2.41 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr20_-_55841398 | 2.40 |
ENST00000395864.3
|
BMP7
|
bone morphogenetic protein 7 |
| chr11_-_125932685 | 2.34 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr4_-_75719896 | 2.33 |
ENST00000395743.3
|
BTC
|
betacellulin |
| chr14_+_33408449 | 2.29 |
ENST00000346562.2
ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr15_-_71146460 | 2.23 |
ENST00000344870.4
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr20_-_55841662 | 2.17 |
ENST00000395863.3
ENST00000450594.2 |
BMP7
|
bone morphogenetic protein 7 |
| chr3_-_42744312 | 2.12 |
ENST00000416756.1
ENST00000441594.1 |
HHATL
|
hedgehog acyltransferase-like |
| chr16_+_86544113 | 2.12 |
ENST00000262426.4
|
FOXF1
|
forkhead box F1 |
| chr12_+_53443680 | 2.12 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr17_+_70117153 | 2.08 |
ENST00000245479.2
|
SOX9
|
SRY (sex determining region Y)-box 9 |
| chr19_+_55587266 | 2.08 |
ENST00000201647.6
ENST00000540810.1 |
EPS8L1
|
EPS8-like 1 |
| chr7_-_19157248 | 2.08 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr13_+_37247934 | 2.05 |
ENST00000315190.3
|
SERTM1
|
serine-rich and transmembrane domain containing 1 |
| chr11_-_119211525 | 2.05 |
ENST00000528368.1
|
C1QTNF5
|
C1q and tumor necrosis factor related protein 5 |
| chr12_+_53443963 | 2.00 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr1_-_32169920 | 1.99 |
ENST00000373672.3
ENST00000373668.3 |
COL16A1
|
collagen, type XVI, alpha 1 |
| chr10_-_118502070 | 1.96 |
ENST00000369209.3
|
HSPA12A
|
heat shock 70kDa protein 12A |
| chr3_+_112930306 | 1.96 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr2_+_74120094 | 1.96 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr4_+_55524085 | 1.90 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr20_+_19193269 | 1.89 |
ENST00000328041.6
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr17_+_60704762 | 1.84 |
ENST00000303375.5
|
MRC2
|
mannose receptor, C type 2 |
| chr9_+_109625378 | 1.84 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr17_+_77020224 | 1.84 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr2_-_27718052 | 1.83 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr10_+_18429606 | 1.83 |
ENST00000324631.7
ENST00000352115.6 ENST00000377328.1 |
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr1_+_160085501 | 1.82 |
ENST00000361216.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr15_-_71146347 | 1.79 |
ENST00000559140.2
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr15_-_63674034 | 1.79 |
ENST00000344366.3
ENST00000422263.2 |
CA12
|
carbonic anhydrase XII |
| chr1_+_160085567 | 1.78 |
ENST00000392233.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr19_-_54984354 | 1.78 |
ENST00000301200.2
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| chr1_-_153585539 | 1.74 |
ENST00000368706.4
|
S100A16
|
S100 calcium binding protein A16 |
| chr5_+_173472607 | 1.74 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr1_+_209859510 | 1.74 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr9_-_35689900 | 1.74 |
ENST00000378300.5
ENST00000329305.2 ENST00000360958.2 |
TPM2
|
tropomyosin 2 (beta) |
| chrX_+_103031758 | 1.74 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr6_-_31514516 | 1.73 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr1_-_32169761 | 1.72 |
ENST00000271069.6
|
COL16A1
|
collagen, type XVI, alpha 1 |
| chr1_+_205473865 | 1.72 |
ENST00000506215.1
ENST00000419301.1 |
CDK18
|
cyclin-dependent kinase 18 |
| chrX_+_103031421 | 1.70 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr4_+_157997273 | 1.69 |
ENST00000541722.1
ENST00000512619.1 |
GLRB
|
glycine receptor, beta |
| chr17_+_77020325 | 1.69 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr6_-_32077100 | 1.68 |
ENST00000375244.3
ENST00000375247.2 |
TNXB
|
tenascin XB |
| chr3_-_42744270 | 1.67 |
ENST00000457462.1
|
HHATL
|
hedgehog acyltransferase-like |
| chr19_+_48216600 | 1.66 |
ENST00000263277.3
ENST00000538399.1 |
EHD2
|
EH-domain containing 2 |
| chrX_-_21776281 | 1.65 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr12_-_91573249 | 1.64 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr10_+_18429671 | 1.61 |
ENST00000282343.8
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr22_+_29876197 | 1.59 |
ENST00000310624.6
|
NEFH
|
neurofilament, heavy polypeptide |
| chr9_-_129884902 | 1.58 |
ENST00000373417.1
|
ANGPTL2
|
angiopoietin-like 2 |
| chr14_-_23834411 | 1.58 |
ENST00000429593.2
|
EFS
|
embryonal Fyn-associated substrate |
| chr19_-_35626104 | 1.57 |
ENST00000310123.3
ENST00000392225.3 |
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chrX_-_110039038 | 1.57 |
ENST00000372042.1
ENST00000482160.1 ENST00000444321.2 ENST00000218054.4 |
CHRDL1
|
chordin-like 1 |
| chr2_+_39893043 | 1.57 |
ENST00000281961.2
|
TMEM178A
|
transmembrane protein 178A |
| chr11_+_125034586 | 1.56 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr9_-_129885010 | 1.56 |
ENST00000373425.3
|
ANGPTL2
|
angiopoietin-like 2 |
| chr1_-_168106536 | 1.56 |
ENST00000537209.1
ENST00000361697.2 ENST00000546300.1 ENST00000367835.1 |
GPR161
|
G protein-coupled receptor 161 |
| chr19_-_51071302 | 1.54 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr7_+_153749732 | 1.53 |
ENST00000377770.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr19_-_4559814 | 1.53 |
ENST00000586582.1
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr1_+_224803995 | 1.53 |
ENST00000272133.3
|
CNIH3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr9_-_112083229 | 1.52 |
ENST00000374566.3
ENST00000374557.4 |
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B |
| chr14_+_53019822 | 1.51 |
ENST00000321662.6
|
GPR137C
|
G protein-coupled receptor 137C |
| chr12_-_91573132 | 1.51 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr21_+_17961006 | 1.50 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chrX_+_70364667 | 1.50 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr10_+_119301928 | 1.49 |
ENST00000553456.3
|
EMX2
|
empty spiracles homeobox 2 |
| chr12_+_119616447 | 1.48 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr6_-_31514333 | 1.46 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr6_+_150464155 | 1.46 |
ENST00000361131.4
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr4_+_62066941 | 1.43 |
ENST00000512091.2
|
LPHN3
|
latrophilin 3 |
| chr3_+_35681081 | 1.43 |
ENST00000428373.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_-_51259528 | 1.43 |
ENST00000404971.1
|
NRXN1
|
neurexin 1 |
| chr1_-_23694794 | 1.43 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr4_+_148402069 | 1.42 |
ENST00000358556.4
ENST00000339690.5 ENST00000511804.1 ENST00000324300.5 |
EDNRA
|
endothelin receptor type A |
| chr15_-_37392086 | 1.42 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr18_-_22932080 | 1.41 |
ENST00000584787.1
ENST00000361524.3 ENST00000538137.2 |
ZNF521
|
zinc finger protein 521 |
| chr19_+_3224700 | 1.40 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr19_+_36359341 | 1.40 |
ENST00000221891.4
|
APLP1
|
amyloid beta (A4) precursor-like protein 1 |
| chr17_+_77020146 | 1.40 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_-_44306590 | 1.40 |
ENST00000377950.3
|
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr6_-_169654139 | 1.40 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr1_+_162039558 | 1.39 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr2_-_68547061 | 1.39 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr13_+_39261224 | 1.37 |
ENST00000280481.7
|
FREM2
|
FRAS1 related extracellular matrix protein 2 |
| chr7_-_38670957 | 1.37 |
ENST00000325590.5
ENST00000428293.2 |
AMPH
|
amphiphysin |
| chr19_-_55658281 | 1.36 |
ENST00000585321.2
ENST00000587465.2 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr2_-_51259641 | 1.34 |
ENST00000406316.2
ENST00000405581.1 |
NRXN1
|
neurexin 1 |
| chr5_-_168728103 | 1.33 |
ENST00000519560.1
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr20_-_44455976 | 1.33 |
ENST00000372555.3
|
TNNC2
|
troponin C type 2 (fast) |
| chr12_-_100378006 | 1.32 |
ENST00000547776.2
ENST00000329257.7 ENST00000547010.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_-_110039286 | 1.31 |
ENST00000434224.1
|
CHRDL1
|
chordin-like 1 |
| chr17_+_64961026 | 1.31 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr19_+_35783028 | 1.29 |
ENST00000600291.1
ENST00000392213.3 |
MAG
|
myelin associated glycoprotein |
| chr4_-_44450814 | 1.29 |
ENST00000360029.3
|
KCTD8
|
potassium channel tetramerization domain containing 8 |
| chr5_+_78985673 | 1.28 |
ENST00000446378.2
|
CMYA5
|
cardiomyopathy associated 5 |
| chr11_-_57417405 | 1.28 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr3_+_112930387 | 1.28 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr14_+_95078714 | 1.27 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr1_+_164528616 | 1.27 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr8_-_144923112 | 1.27 |
ENST00000442628.2
|
NRBP2
|
nuclear receptor binding protein 2 |
| chr15_-_63674218 | 1.27 |
ENST00000178638.3
|
CA12
|
carbonic anhydrase XII |
| chr6_+_31949801 | 1.27 |
ENST00000428956.2
ENST00000498271.1 |
C4A
|
complement component 4A (Rodgers blood group) |
| chr19_+_35783047 | 1.26 |
ENST00000595791.1
ENST00000597035.1 ENST00000537831.2 |
MAG
|
myelin associated glycoprotein |
| chr5_+_128301189 | 1.26 |
ENST00000395266.1
ENST00000506176.1 |
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr5_-_11904100 | 1.24 |
ENST00000359640.2
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr4_-_5891918 | 1.23 |
ENST00000512574.1
|
CRMP1
|
collapsin response mediator protein 1 |
| chr12_+_52626898 | 1.23 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr9_+_87284622 | 1.22 |
ENST00000395882.1
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr6_+_31982539 | 1.21 |
ENST00000435363.2
ENST00000425700.2 |
C4B
|
complement component 4B (Chido blood group) |
| chr3_+_112929850 | 1.21 |
ENST00000464546.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr2_-_218867711 | 1.21 |
ENST00000446903.1
|
TNS1
|
tensin 1 |
| chr10_-_125651258 | 1.21 |
ENST00000241305.3
|
CPXM2
|
carboxypeptidase X (M14 family), member 2 |
| chr2_+_220325441 | 1.20 |
ENST00000396688.1
|
SPEG
|
SPEG complex locus |
| chr5_+_128300810 | 1.20 |
ENST00000262462.4
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr3_-_133614597 | 1.20 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr2_-_11810284 | 1.20 |
ENST00000306928.5
|
NTSR2
|
neurotensin receptor 2 |
| chr19_-_47975143 | 1.19 |
ENST00000597014.1
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr9_+_95947198 | 1.18 |
ENST00000448039.1
ENST00000297954.4 ENST00000395477.2 ENST00000395475.2 ENST00000349097.3 ENST00000427277.2 ENST00000356055.3 ENST00000432730.1 |
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr11_+_125034640 | 1.18 |
ENST00000542175.1
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr6_-_32077078 | 1.18 |
ENST00000479795.1
|
TNXB
|
tenascin XB |
| chr16_+_4845379 | 1.17 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr3_-_58522880 | 1.17 |
ENST00000474098.1
|
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr15_+_41851211 | 1.17 |
ENST00000263798.3
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr5_-_11904152 | 1.17 |
ENST00000304623.8
ENST00000458100.2 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr14_+_77564701 | 1.17 |
ENST00000557115.1
|
KIAA1737
|
CLOCK-interacting pacemaker |
| chr6_+_31950150 | 1.16 |
ENST00000537134.1
|
C4A
|
complement component 4A (Rodgers blood group) |
| chr19_-_47157914 | 1.16 |
ENST00000300875.4
|
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr10_+_123872483 | 1.16 |
ENST00000369001.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr19_+_35783037 | 1.16 |
ENST00000361922.4
|
MAG
|
myelin associated glycoprotein |
| chr19_-_55658650 | 1.15 |
ENST00000589226.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chrX_+_70435044 | 1.15 |
ENST00000374029.1
ENST00000374022.3 ENST00000447581.1 |
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr2_-_230579185 | 1.14 |
ENST00000341772.4
|
DNER
|
delta/notch-like EGF repeat containing |
| chr5_+_140782351 | 1.13 |
ENST00000573521.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chr1_-_225840747 | 1.13 |
ENST00000366843.2
ENST00000366844.3 |
ENAH
|
enabled homolog (Drosophila) |
| chrX_-_130423240 | 1.13 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr10_-_79397202 | 1.13 |
ENST00000372437.1
ENST00000372408.2 ENST00000372403.4 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_-_47207390 | 1.13 |
ENST00000539589.1
ENST00000528462.1 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr7_+_100081542 | 1.12 |
ENST00000300179.2
ENST00000423930.1 |
NYAP1
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1 |
| chr3_-_133614421 | 1.12 |
ENST00000543906.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr18_+_33877654 | 1.12 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr12_-_96184533 | 1.12 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr3_-_42744130 | 1.11 |
ENST00000417472.1
ENST00000442469.1 |
HHATL
|
hedgehog acyltransferase-like |
| chr3_-_62861012 | 1.11 |
ENST00000357948.3
ENST00000383710.4 |
CADPS
|
Ca++-dependent secretion activator |
| chr10_-_79397391 | 1.11 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_+_130318869 | 1.09 |
ENST00000299164.2
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr2_-_51259292 | 1.09 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr15_-_83378638 | 1.09 |
ENST00000261722.3
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr7_-_140340576 | 1.09 |
ENST00000275884.6
ENST00000475837.1 |
DENND2A
|
DENN/MADD domain containing 2A |
| chr10_-_79397316 | 1.09 |
ENST00000372421.5
ENST00000457953.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr8_-_75233563 | 1.09 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr20_-_18038521 | 1.08 |
ENST00000278780.6
|
OVOL2
|
ovo-like zinc finger 2 |
| chr7_-_14942467 | 1.08 |
ENST00000407950.1
ENST00000444700.2 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr12_+_6419877 | 1.07 |
ENST00000536531.1
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr4_-_77819002 | 1.07 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr1_-_153588765 | 1.07 |
ENST00000368701.1
ENST00000344616.2 |
S100A14
|
S100 calcium binding protein A14 |
| chr11_-_57004658 | 1.07 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr16_-_28550320 | 1.07 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr1_+_205473784 | 1.06 |
ENST00000478560.1
ENST00000443813.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr5_+_32710736 | 1.06 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_-_153588334 | 1.06 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14 |
| chr3_-_120169828 | 1.06 |
ENST00000424703.2
ENST00000469005.1 |
FSTL1
|
follistatin-like 1 |
| chr20_-_42816206 | 1.06 |
ENST00000372980.3
|
JPH2
|
junctophilin 2 |
| chr9_+_87284675 | 1.05 |
ENST00000376208.1
ENST00000304053.6 ENST00000277120.3 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr12_-_91573316 | 1.04 |
ENST00000393155.1
|
DCN
|
decorin |
| chr11_+_60691924 | 1.04 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr19_-_49339080 | 1.04 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr7_-_51384451 | 1.04 |
ENST00000441453.1
ENST00000265136.7 ENST00000395542.2 ENST00000395540.2 |
COBL
|
cordon-bleu WH2 repeat protein |
| chr2_+_18059906 | 1.04 |
ENST00000304101.4
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr9_-_122131696 | 1.03 |
ENST00000373964.2
ENST00000265922.3 |
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chrX_+_24167828 | 1.03 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr19_-_19373601 | 1.03 |
ENST00000291481.7
|
HAPLN4
|
hyaluronan and proteoglycan link protein 4 |
| chr1_+_150480551 | 1.03 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr10_+_73724123 | 1.03 |
ENST00000373115.4
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr7_+_32996997 | 1.03 |
ENST00000242209.4
ENST00000538336.1 ENST00000538443.1 |
FKBP9
|
FK506 binding protein 9, 63 kDa |
| chr11_+_73019282 | 1.02 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr1_+_155290677 | 1.02 |
ENST00000368354.3
ENST00000368352.5 |
RUSC1
|
RUN and SH3 domain containing 1 |
| chr7_+_94023873 | 1.02 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr6_+_44238203 | 1.02 |
ENST00000451188.2
|
TMEM151B
|
transmembrane protein 151B |
| chr2_-_68547019 | 1.02 |
ENST00000409862.1
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr8_+_19796381 | 1.01 |
ENST00000524029.1
ENST00000522701.1 ENST00000311322.8 |
LPL
|
lipoprotein lipase |
| chr21_-_40984704 | 1.01 |
ENST00000329618.6
|
C21orf88
|
chromosome 21 open reading frame 88 |
| chr13_-_110959478 | 1.01 |
ENST00000543140.1
ENST00000375820.4 |
COL4A1
|
collagen, type IV, alpha 1 |
| chr19_-_49243845 | 1.01 |
ENST00000222145.4
|
RASIP1
|
Ras interacting protein 1 |
| chr16_+_66878282 | 1.01 |
ENST00000338437.2
|
CA7
|
carbonic anhydrase VII |
| chr19_-_51466681 | 1.01 |
ENST00000456750.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chrX_-_72095808 | 1.00 |
ENST00000373529.5
|
DMRTC1
|
DMRT-like family C1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF263
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 2.1 | GO:0060516 | primary prostatic bud elongation(GO:0060516) |
| 1.2 | 4.9 | GO:0060254 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 1.2 | 3.6 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 1.1 | 4.6 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) allantois development(GO:1905069) |
| 1.0 | 2.1 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 1.0 | 9.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.9 | 2.8 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.9 | 0.9 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.7 | 2.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) right lung development(GO:0060458) |
| 0.6 | 1.9 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.6 | 4.9 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.6 | 2.9 | GO:0048749 | compound eye development(GO:0048749) |
| 0.6 | 1.7 | GO:0002585 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.6 | 5.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.6 | 3.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.5 | 1.6 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.5 | 6.3 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.5 | 1.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.5 | 1.5 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.5 | 1.4 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.5 | 1.4 | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.5 | 1.4 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.5 | 1.8 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.4 | 0.4 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.4 | 3.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 1.2 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.4 | 2.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 3.6 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.4 | 2.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.4 | 2.3 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.4 | 2.3 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.4 | 3.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.4 | 2.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.3 | 3.8 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.3 | 1.0 | GO:0050894 | determination of affect(GO:0050894) |
| 0.3 | 1.7 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.3 | 1.3 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.3 | 1.0 | GO:0086055 | atrial ventricular junction remodeling(GO:0003294) atrial cardiac muscle cell to AV node cell communication by electrical coupling(GO:0086044) bundle of His cell to Purkinje myocyte communication by electrical coupling(GO:0086054) Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling(GO:0086055) regulation of Purkinje myocyte action potential(GO:0098906) vasomotion(GO:1990029) |
| 0.3 | 1.0 | GO:0048925 | lateral line system development(GO:0048925) |
| 0.3 | 1.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 0.9 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.3 | 7.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 1.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.3 | 0.8 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.3 | 0.8 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.3 | 1.4 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.3 | 4.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 1.9 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 | 1.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 3.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 1.0 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.3 | 2.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 0.7 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.2 | 1.7 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 0.7 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.2 | 1.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 6.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 0.9 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.2 | 0.9 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 0.7 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 0.9 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 1.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.2 | 1.0 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.2 | 0.8 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.2 | 2.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 0.4 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.2 | 0.4 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 1.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.2 | 0.8 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.2 | 0.6 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 1.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 7.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.6 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.2 | 0.6 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.2 | 2.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 0.5 | GO:0090274 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 1.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.2 | 0.2 | GO:0043132 | NAD transport(GO:0043132) |
| 0.2 | 3.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 0.7 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.2 | 4.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 1.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 0.5 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.2 | 0.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 0.8 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.2 | 0.5 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.2 | 1.5 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 0.5 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.2 | 1.0 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 1.0 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.2 | 0.5 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.2 | 0.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 1.5 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.2 | 0.6 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 0.6 | GO:0048377 | lateral mesodermal cell differentiation(GO:0048371) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.1 | 0.4 | GO:0060718 | Spemann organizer formation(GO:0060061) chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 1.0 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.1 | 0.7 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 1.9 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) |
| 0.1 | 4.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 2.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 4.0 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 2.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.7 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.5 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.6 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.8 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.2 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 2.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.5 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 3.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.9 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 1.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.6 | GO:0072098 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.1 | 0.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 4.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.5 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 1.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.7 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.9 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 1.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 1.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.3 | GO:0060584 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.3 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.1 | 1.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.7 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.3 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 2.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 1.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.3 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.7 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.8 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.3 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 0.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.3 | GO:0061075 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 2.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.6 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 2.0 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 1.1 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.1 | 0.9 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 1.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 2.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.4 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 3.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.4 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.7 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.9 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.1 | 1.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 1.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 2.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.5 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.6 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 1.8 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 1.7 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.3 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.7 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 0.4 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.1 | 0.6 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 3.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.7 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.6 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 1.3 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 1.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 2.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.6 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.8 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.2 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.5 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 1.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.3 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 0.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 1.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 3.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.7 | GO:0015820 | leucine transport(GO:0015820) |
| 0.1 | 0.5 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 1.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.3 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 1.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 1.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 2.7 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.4 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.1 | 0.3 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 1.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 1.1 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.6 | GO:0030002 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) |
| 0.1 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.3 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.1 | 0.5 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.1 | 1.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.6 | GO:0070779 | sulfur amino acid transport(GO:0000101) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.1 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 2.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.5 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 0.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.6 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.6 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.5 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.5 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 1.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 1.1 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.5 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 1.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.7 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.4 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.8 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 2.5 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 1.7 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.6 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 1.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.9 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 1.4 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.3 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.6 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 1.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.4 | GO:0044557 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 2.2 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 1.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.8 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 3.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 2.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.5 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.6 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.5 | GO:0030815 | negative regulation of cAMP metabolic process(GO:0030815) |
| 0.0 | 1.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.6 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.6 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 1.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.0 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.5 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.3 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 1.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.5 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0072144 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.9 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.5 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.3 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.9 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.5 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.4 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.2 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0000726 | non-recombinational repair(GO:0000726) double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of Rho protein signal transduction(GO:0035025) positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.5 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.4 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.9 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.3 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 1.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.3 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:1902074 | response to salt(GO:1902074) |
| 0.0 | 0.2 | GO:0032680 | regulation of tumor necrosis factor production(GO:0032680) regulation of tumor necrosis factor superfamily cytokine production(GO:1903555) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.0 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.6 | 1.9 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.6 | 1.7 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.4 | 1.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 4.3 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.3 | 2.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.3 | 4.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 1.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 5.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.3 | 0.9 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.3 | 3.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 4.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 1.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 0.8 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.2 | 4.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 5.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.4 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 0.7 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.2 | 0.9 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.2 | 0.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 1.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 3.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 1.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.5 | GO:0034686 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 1.0 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 2.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.8 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 1.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 3.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 4.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.5 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.1 | 0.3 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 1.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 3.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 2.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 12.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 4.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 6.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 1.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 8.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 2.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.1 | 1.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 10.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 6.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.2 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.1 | 9.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 3.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 7.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 3.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.4 | GO:1990351 | transmembrane transporter complex(GO:1902495) transporter complex(GO:1990351) |
| 0.0 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 5.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.8 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 8.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 5.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 10.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 7.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.9 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 5.0 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.4 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.8 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 2.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 3.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.9 | GO:0043296 | apical junction complex(GO:0043296) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.6 | 1.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.6 | 2.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.5 | 2.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 7.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.5 | 8.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 1.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.5 | 4.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 2.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.4 | 1.5 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.4 | 5.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 1.0 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.3 | 6.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 1.7 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.3 | 2.3 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 2.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 1.0 | GO:0086079 | gap junction channel activity involved in atrial cardiac muscle cell-AV node cell electrical coupling(GO:0086076) gap junction channel activity involved in bundle of His cell-Purkinje myocyte electrical coupling(GO:0086078) gap junction channel activity involved in Purkinje myocyte-ventricular cardiac muscle cell electrical coupling(GO:0086079) |
| 0.3 | 4.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 2.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 5.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 0.9 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.3 | 1.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 1.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 2.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 1.3 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.2 | 1.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 2.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.0 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.2 | 3.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.7 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.2 | 1.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 2.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 0.9 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 0.7 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.2 | 0.7 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 0.6 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.2 | 4.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 1.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.2 | 3.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 2.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 1.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 3.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.2 | 3.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 2.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 0.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 1.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.3 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.4 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 1.0 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.7 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 3.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 3.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 2.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.8 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 2.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.5 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.4 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.9 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 1.0 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.1 | 1.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.4 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.5 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.7 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.7 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 2.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 13.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.3 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 1.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.9 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 6.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.6 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.8 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.9 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 2.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.6 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 3.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.3 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 2.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.5 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 1.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.3 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.8 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.6 | GO:0015501 | sulfur amino acid transmembrane transporter activity(GO:0000099) glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.2 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.7 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.7 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.9 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.6 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 1.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 1.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 3.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 1.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 1.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.0 | 3.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 2.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 1.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 2.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 5.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 5.2 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 2.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 1.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 4.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) sequence-specific single stranded DNA binding(GO:0098847) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 10.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 8.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 8.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 16.5 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 10.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 3.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 3.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.3 | 0.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 4.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 7.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 7.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 13.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 4.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 7.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 4.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 5.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 6.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 3.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 3.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 3.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 2.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 2.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 1.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.6 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |