Project
Illumina Body Map 2
Navigation
Downloads
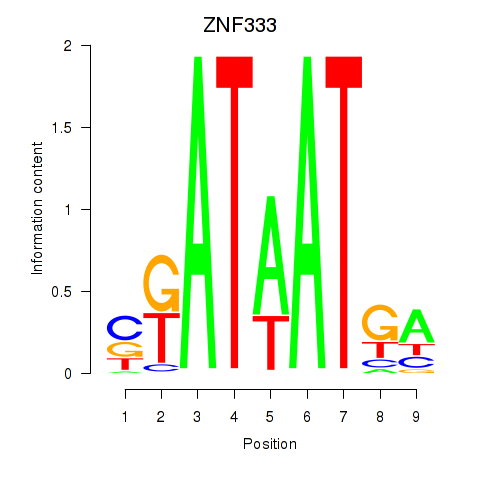
Results for ZNF333
Z-value: 0.98
Transcription factors associated with ZNF333
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF333
|
ENSG00000160961.7 | zinc finger protein 333 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF333 | hg19_v2_chr19_+_14800711_14800917 | 0.17 | 3.6e-01 | Click! |
Activity profile of ZNF333 motif
Sorted Z-values of ZNF333 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_35721182 | 3.80 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chrX_-_21776281 | 3.22 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr10_+_50507232 | 3.10 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr5_+_137203465 | 2.87 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr12_-_49393092 | 2.84 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr10_+_50507181 | 2.75 |
ENST00000323868.4
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr4_-_177190364 | 2.69 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr3_+_35721106 | 2.50 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_35721130 | 2.23 |
ENST00000432450.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr12_+_122880045 | 1.56 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr3_+_14716606 | 1.40 |
ENST00000253697.3
ENST00000435614.1 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr14_+_23709555 | 1.37 |
ENST00000430154.2
|
C14orf164
|
chromosome 14 open reading frame 164 |
| chrX_-_110507098 | 1.24 |
ENST00000541758.1
|
CAPN6
|
calpain 6 |
| chr15_+_86686953 | 1.21 |
ENST00000421325.2
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr10_-_128210005 | 1.20 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr6_+_50786414 | 1.20 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr13_-_114107839 | 1.16 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr2_-_178753465 | 1.03 |
ENST00000389683.3
|
PDE11A
|
phosphodiesterase 11A |
| chr15_+_54305101 | 1.02 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr19_-_19626838 | 1.01 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr19_-_19626351 | 0.97 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr11_-_26744908 | 0.97 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr9_-_7961080 | 0.96 |
ENST00000435444.1
|
RP11-29B9.2
|
RP11-29B9.2 |
| chrX_-_49121165 | 0.93 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr1_-_11024258 | 0.90 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr6_+_35748783 | 0.88 |
ENST00000373861.5
ENST00000542261.1 |
CLPSL1
|
colipase-like 1 |
| chr14_+_22992573 | 0.85 |
ENST00000390516.1
|
TRAJ21
|
T cell receptor alpha joining 21 |
| chr12_-_7848364 | 0.83 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr1_+_81106951 | 0.79 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr9_+_109625378 | 0.78 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr10_+_121410882 | 0.76 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr10_+_90346519 | 0.75 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr15_+_36338242 | 0.75 |
ENST00000560056.1
|
RP11-684B21.1
|
RP11-684B21.1 |
| chr4_+_66536248 | 0.74 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr5_-_54988559 | 0.73 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr12_-_87232644 | 0.73 |
ENST00000549405.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_-_42360015 | 0.70 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr7_-_142240014 | 0.67 |
ENST00000390363.2
|
TRBV9
|
T cell receptor beta variable 9 |
| chr15_+_22368478 | 0.67 |
ENST00000332663.2
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2 |
| chr1_+_36789335 | 0.67 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr6_-_32908765 | 0.65 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr5_-_54988448 | 0.64 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr12_+_58176525 | 0.64 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr5_+_169011033 | 0.63 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr5_-_16509101 | 0.63 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr6_-_32908792 | 0.62 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr1_-_205391178 | 0.62 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr6_-_2634820 | 0.61 |
ENST00000296847.3
|
C6orf195
|
chromosome 6 open reading frame 195 |
| chr4_+_71019903 | 0.60 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr3_-_112564797 | 0.58 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr20_+_56964169 | 0.57 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr11_+_18433840 | 0.57 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr12_-_122879969 | 0.56 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr3_+_38323785 | 0.56 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr13_-_103719196 | 0.56 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr1_-_51763661 | 0.53 |
ENST00000530004.1
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr2_+_191002486 | 0.53 |
ENST00000396974.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr1_-_115301235 | 0.49 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr20_+_59654146 | 0.49 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr14_+_20344391 | 0.49 |
ENST00000298642.2
|
OR4K2
|
olfactory receptor, family 4, subfamily K, member 2 |
| chr11_-_117698787 | 0.48 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr1_+_19638788 | 0.47 |
ENST00000375155.3
ENST00000375153.3 ENST00000400548.2 |
PQLC2
|
PQ loop repeat containing 2 |
| chr12_-_52995210 | 0.45 |
ENST00000398066.3
|
KRT72
|
keratin 72 |
| chr11_-_117698765 | 0.45 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr9_+_133971909 | 0.45 |
ENST00000247291.3
ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1-like |
| chr12_-_52995242 | 0.44 |
ENST00000537672.2
|
KRT72
|
keratin 72 |
| chr5_+_169010638 | 0.44 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr4_+_71108300 | 0.43 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr19_-_28957557 | 0.43 |
ENST00000585697.1
ENST00000591920.1 |
AC005307.3
|
AC005307.3 |
| chr5_-_9630463 | 0.43 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr11_-_123814580 | 0.42 |
ENST00000321252.2
|
OR6T1
|
olfactory receptor, family 6, subfamily T, member 1 |
| chr11_-_55371874 | 0.42 |
ENST00000302231.4
|
OR4C11
|
olfactory receptor, family 4, subfamily C, member 11 |
| chr15_+_99433570 | 0.42 |
ENST00000558898.1
|
IGF1R
|
insulin-like growth factor 1 receptor |
| chr20_+_56964253 | 0.41 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr2_-_235363231 | 0.41 |
ENST00000439798.1
|
AC097713.4
|
AC097713.4 |
| chr12_-_52995291 | 0.41 |
ENST00000293745.2
ENST00000354310.4 |
KRT72
|
keratin 72 |
| chr9_+_133971863 | 0.41 |
ENST00000372309.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr1_-_47131521 | 0.40 |
ENST00000542495.1
ENST00000532925.1 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr14_+_85996471 | 0.40 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr11_+_94300474 | 0.40 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr17_-_46690839 | 0.38 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr1_+_159409512 | 0.38 |
ENST00000423932.3
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1 |
| chr10_+_118350468 | 0.38 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr6_+_76330355 | 0.37 |
ENST00000483859.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr9_-_133769225 | 0.37 |
ENST00000343079.1
|
QRFP
|
pyroglutamylated RFamide peptide |
| chr8_+_92114060 | 0.35 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr15_-_88247083 | 0.35 |
ENST00000560439.1
|
RP11-648K4.2
|
RP11-648K4.2 |
| chr1_+_109265033 | 0.35 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr2_+_79412357 | 0.33 |
ENST00000466387.1
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr17_-_33390667 | 0.33 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr11_-_4389616 | 0.32 |
ENST00000408920.2
|
OR52B4
|
olfactory receptor, family 52, subfamily B, member 4 |
| chr17_-_71223839 | 0.32 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chrX_+_47867194 | 0.32 |
ENST00000376940.3
|
SPACA5
|
sperm acrosome associated 5 |
| chr10_+_118349920 | 0.32 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr1_-_198990166 | 0.31 |
ENST00000427439.1
|
RP11-16L9.3
|
RP11-16L9.3 |
| chr5_-_27038683 | 0.31 |
ENST00000511822.1
ENST00000231021.4 |
CDH9
|
cadherin 9, type 2 (T1-cadherin) |
| chr3_-_126327398 | 0.31 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr5_-_35195338 | 0.31 |
ENST00000509839.1
|
PRLR
|
prolactin receptor |
| chr5_+_137722255 | 0.31 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr1_+_218458625 | 0.31 |
ENST00000366932.3
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae) |
| chr11_-_71823796 | 0.31 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chrX_+_47990039 | 0.31 |
ENST00000304270.5
|
SPACA5B
|
sperm acrosome associated 5B |
| chr3_-_37216055 | 0.31 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr14_+_20248401 | 0.31 |
ENST00000315957.4
|
OR4M1
|
olfactory receptor, family 4, subfamily M, member 1 |
| chr5_+_173315283 | 0.30 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr8_-_134114887 | 0.30 |
ENST00000519341.1
|
SLA
|
Src-like-adaptor |
| chr14_+_85996507 | 0.29 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr7_+_1272522 | 0.29 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr9_-_21351377 | 0.29 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr1_-_36020531 | 0.29 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr6_+_29274403 | 0.28 |
ENST00000377160.2
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1 |
| chr7_-_105319536 | 0.28 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr5_+_137225125 | 0.28 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr7_-_130080977 | 0.27 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr4_+_54927213 | 0.27 |
ENST00000595906.1
|
AC110792.1
|
HCG2027126; Uncharacterized protein |
| chr12_-_10959892 | 0.27 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chr21_-_39705286 | 0.26 |
ENST00000414189.1
|
AP001422.3
|
AP001422.3 |
| chr16_+_53412368 | 0.26 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr11_+_10772534 | 0.26 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr20_+_44330651 | 0.26 |
ENST00000305479.2
|
WFDC13
|
WAP four-disulfide core domain 13 |
| chr10_+_118350522 | 0.26 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr10_-_69425399 | 0.26 |
ENST00000330298.6
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr17_-_38911580 | 0.26 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chrX_+_11311533 | 0.25 |
ENST00000380714.3
ENST00000380712.3 ENST00000348912.4 |
AMELX
|
amelogenin, X-linked |
| chr20_+_58515417 | 0.24 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr5_+_137225158 | 0.24 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr14_+_105046094 | 0.24 |
ENST00000331952.2
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr3_-_125094093 | 0.24 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr20_+_1099233 | 0.23 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr3_-_196295385 | 0.23 |
ENST00000425888.1
|
WDR53
|
WD repeat domain 53 |
| chr6_-_52705641 | 0.23 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr12_+_109785708 | 0.23 |
ENST00000310903.5
|
MYO1H
|
myosin IH |
| chr10_-_32667660 | 0.22 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr20_-_35329063 | 0.22 |
ENST00000422536.1
|
NDRG3
|
NDRG family member 3 |
| chr2_-_228497888 | 0.22 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr13_-_30160925 | 0.22 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr10_-_23633720 | 0.21 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr18_+_71815743 | 0.21 |
ENST00000169551.6
ENST00000580087.1 |
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr16_-_28222797 | 0.21 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr20_+_22201070 | 0.20 |
ENST00000439450.1
|
RP5-1097P24.1
|
RP5-1097P24.1 |
| chr14_+_23842018 | 0.20 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr11_-_18062872 | 0.20 |
ENST00000250018.2
|
TPH1
|
tryptophan hydroxylase 1 |
| chr2_-_27435390 | 0.20 |
ENST00000428518.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chrX_+_99839799 | 0.19 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr11_-_71823715 | 0.19 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr3_-_113918254 | 0.19 |
ENST00000460779.1
|
DRD3
|
dopamine receptor D3 |
| chr19_-_45004556 | 0.19 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr8_+_17013515 | 0.18 |
ENST00000262096.8
|
ZDHHC2
|
zinc finger, DHHC-type containing 2 |
| chr15_-_96831660 | 0.18 |
ENST00000378936.1
|
AC016251.1
|
AC016251.1 |
| chr20_-_44600810 | 0.18 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr1_+_215179188 | 0.17 |
ENST00000391895.2
|
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr14_+_20403767 | 0.17 |
ENST00000285600.4
|
OR4K1
|
olfactory receptor, family 4, subfamily K, member 1 |
| chr1_+_247670360 | 0.17 |
ENST00000527084.1
ENST00000527541.1 ENST00000366490.3 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chr9_+_22646189 | 0.17 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr8_-_11725549 | 0.16 |
ENST00000505496.2
ENST00000534636.1 ENST00000524500.1 ENST00000345125.3 ENST00000453527.2 ENST00000527215.2 ENST00000532392.1 ENST00000533455.1 ENST00000534510.1 ENST00000530640.2 ENST00000531089.1 ENST00000532656.2 ENST00000531502.1 ENST00000434271.1 ENST00000353047.6 |
CTSB
|
cathepsin B |
| chr1_+_248112160 | 0.16 |
ENST00000357191.3
|
OR2L8
|
olfactory receptor, family 2, subfamily L, member 8 (gene/pseudogene) |
| chr9_-_19786926 | 0.16 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr16_+_20499024 | 0.15 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr2_-_151905288 | 0.14 |
ENST00000409243.1
|
AC023469.1
|
HCG1817310; Uncharacterized protein |
| chr1_+_55013889 | 0.14 |
ENST00000343744.2
ENST00000371316.3 |
ACOT11
|
acyl-CoA thioesterase 11 |
| chr10_-_4720333 | 0.13 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr10_+_54074033 | 0.12 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr3_-_194393206 | 0.11 |
ENST00000265245.5
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr7_+_141463897 | 0.11 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr2_+_233527443 | 0.11 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr10_-_128975273 | 0.11 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr8_+_21823726 | 0.11 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr22_-_32651326 | 0.11 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr14_+_76618242 | 0.10 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chrM_+_10053 | 0.10 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr11_-_66313699 | 0.10 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr8_+_61429416 | 0.10 |
ENST00000262646.7
ENST00000531289.1 |
RAB2A
|
RAB2A, member RAS oncogene family |
| chr11_-_58275578 | 0.10 |
ENST00000360374.2
|
OR5B21
|
olfactory receptor, family 5, subfamily B, member 21 |
| chr12_-_75905374 | 0.09 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr19_+_13122980 | 0.09 |
ENST00000590027.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr11_-_6173818 | 0.09 |
ENST00000316506.1
|
OR52B1P
|
olfactory receptor, family 52, subfamily B, member 1 pseudogene |
| chr12_+_92817735 | 0.09 |
ENST00000378485.1
|
CLLU1
|
chronic lymphocytic leukemia up-regulated 1 |
| chr4_+_146402346 | 0.09 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr8_-_134114721 | 0.09 |
ENST00000522119.1
ENST00000523610.1 ENST00000521302.1 ENST00000519558.1 ENST00000519747.1 ENST00000517648.1 |
SLA
|
Src-like-adaptor |
| chrY_+_14958970 | 0.09 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr5_-_95018660 | 0.09 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr4_+_110749143 | 0.08 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr21_-_46964306 | 0.08 |
ENST00000443742.1
ENST00000528477.1 ENST00000567670.1 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr11_+_4615037 | 0.07 |
ENST00000450052.2
|
OR52I1
|
olfactory receptor, family 52, subfamily I, member 1 |
| chr20_-_58515344 | 0.07 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chr14_+_105046021 | 0.06 |
ENST00000557649.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr20_-_33413416 | 0.06 |
ENST00000359003.2
|
NCOA6
|
nuclear receptor coactivator 6 |
| chr19_+_19626531 | 0.06 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr5_-_78281623 | 0.06 |
ENST00000521117.1
|
ARSB
|
arylsulfatase B |
| chr5_+_140535577 | 0.05 |
ENST00000539533.1
|
PCDHB17
|
Protocadherin-psi1; Uncharacterized protein |
| chr12_-_100656134 | 0.05 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr11_-_7961141 | 0.05 |
ENST00000360759.3
|
OR10A3
|
olfactory receptor, family 10, subfamily A, member 3 |
| chr8_+_107460147 | 0.04 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chr17_-_33446735 | 0.04 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr7_+_6797288 | 0.04 |
ENST00000433859.2
ENST00000359718.3 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr12_-_7656357 | 0.04 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr7_-_36406750 | 0.04 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr5_-_78281775 | 0.04 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr7_+_6796986 | 0.03 |
ENST00000297186.3
|
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr5_+_38403637 | 0.03 |
ENST00000336740.6
ENST00000397202.2 |
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF333
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.9 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.4 | 1.3 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.2 | 0.6 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.2 | 1.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.4 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 2.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.7 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 1.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.2 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.1 | 0.8 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.7 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 1.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.3 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.2 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 1.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 1.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0100012 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.2 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.6 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 1.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 6.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.4 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.3 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.7 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.9 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 3.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.3 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.1 | 2.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.6 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 1.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.4 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 1.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.7 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 3.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 1.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.6 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 8.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 3.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |