Project
Illumina Body Map 2
Navigation
Downloads
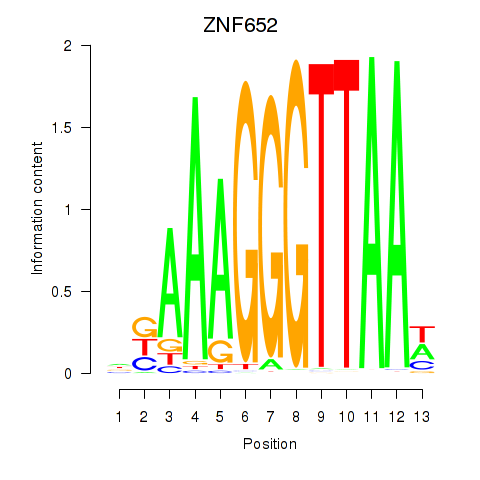
Results for ZNF652
Z-value: 1.49
Transcription factors associated with ZNF652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF652
|
ENSG00000198740.4 | zinc finger protein 652 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF652 | hg19_v2_chr17_-_47439437_47439533 | 0.13 | 4.8e-01 | Click! |
Activity profile of ZNF652 motif
Sorted Z-values of ZNF652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_90757364 | 5.22 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_90756769 | 4.35 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chrX_+_103031758 | 4.24 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr17_-_10741762 | 3.76 |
ENST00000580256.2
|
PIRT
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr2_-_220174166 | 3.71 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr11_-_130298888 | 3.63 |
ENST00000257359.6
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr6_-_34524049 | 3.34 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr6_-_34524093 | 3.15 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr9_+_74764340 | 3.12 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr16_+_56623433 | 3.11 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr10_-_10836919 | 2.94 |
ENST00000602763.1
ENST00000415590.2 ENST00000434919.2 |
SFTA1P
|
surfactant associated 1, pseudogene |
| chr7_-_140341251 | 2.92 |
ENST00000491728.1
|
DENND2A
|
DENN/MADD domain containing 2A |
| chr11_-_82444892 | 2.77 |
ENST00000329203.3
|
FAM181B
|
family with sequence similarity 181, member B |
| chr11_-_35547572 | 2.73 |
ENST00000378880.2
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr2_+_11052054 | 2.68 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr9_-_117853297 | 2.64 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr14_+_79746249 | 2.63 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr19_-_51472031 | 2.60 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chrX_-_128788914 | 2.60 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr11_-_35547151 | 2.56 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr11_-_35547277 | 2.48 |
ENST00000527605.1
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr9_+_74764278 | 2.39 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr5_-_45696253 | 2.33 |
ENST00000303230.4
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr1_+_86934526 | 2.32 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr5_-_153857819 | 2.27 |
ENST00000231121.2
|
HAND1
|
heart and neural crest derivatives expressed 1 |
| chr17_-_77179487 | 2.26 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr17_+_48610074 | 2.26 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr11_-_111782696 | 2.26 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr5_+_161494521 | 2.23 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chrX_-_20074895 | 2.23 |
ENST00000543767.1
|
MAP7D2
|
MAP7 domain containing 2 |
| chr5_+_161494770 | 2.20 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr14_+_79745682 | 2.18 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr14_+_79745746 | 2.14 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr16_+_24266874 | 2.05 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr19_+_11649532 | 2.05 |
ENST00000252456.2
ENST00000592923.1 ENST00000535659.2 |
CNN1
|
calponin 1, basic, smooth muscle |
| chr11_-_111782484 | 2.02 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr17_+_47653471 | 1.96 |
ENST00000513748.1
|
NXPH3
|
neurexophilin 3 |
| chr11_-_119293872 | 1.94 |
ENST00000524970.1
|
THY1
|
Thy-1 cell surface antigen |
| chr12_+_20848377 | 1.90 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr8_-_110656995 | 1.81 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr14_-_22005343 | 1.80 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr3_+_157154578 | 1.79 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr5_-_176057518 | 1.77 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr10_-_10836865 | 1.76 |
ENST00000446372.2
|
SFTA1P
|
surfactant associated 1, pseudogene |
| chr12_-_120241187 | 1.74 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr16_-_57318566 | 1.73 |
ENST00000569059.1
ENST00000219207.5 |
PLLP
|
plasmolipin |
| chr3_+_183948161 | 1.72 |
ENST00000426955.2
|
VWA5B2
|
von Willebrand factor A domain containing 5B2 |
| chr19_-_47137942 | 1.70 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr17_-_46035187 | 1.63 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr5_-_176057365 | 1.62 |
ENST00000310112.3
|
SNCB
|
synuclein, beta |
| chr7_-_140340576 | 1.59 |
ENST00000275884.6
ENST00000475837.1 |
DENND2A
|
DENN/MADD domain containing 2A |
| chr14_-_22005062 | 1.59 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr12_-_6809958 | 1.57 |
ENST00000320591.5
ENST00000534837.1 |
PIANP
|
PILR alpha associated neural protein |
| chr7_-_142583506 | 1.56 |
ENST00000359396.3
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr1_+_162601774 | 1.55 |
ENST00000415555.1
|
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr5_+_152870287 | 1.54 |
ENST00000340592.5
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr1_-_156675564 | 1.54 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_+_48610042 | 1.52 |
ENST00000503246.1
|
EPN3
|
epsin 3 |
| chr17_+_38337491 | 1.52 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr4_+_81118647 | 1.48 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr17_+_26800296 | 1.47 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr19_+_55795493 | 1.43 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr1_+_26348259 | 1.42 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr10_-_100995603 | 1.42 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
| chr12_+_113229962 | 1.40 |
ENST00000553114.1
ENST00000548866.1 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr1_-_156647189 | 1.40 |
ENST00000368223.3
|
NES
|
nestin |
| chr6_+_72926145 | 1.38 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr19_-_51143075 | 1.38 |
ENST00000600079.1
ENST00000593901.1 |
SYT3
|
synaptotagmin III |
| chr17_-_45928521 | 1.37 |
ENST00000536300.1
|
SP6
|
Sp6 transcription factor |
| chr19_-_51142540 | 1.37 |
ENST00000598997.1
|
SYT3
|
synaptotagmin III |
| chr9_+_131174024 | 1.37 |
ENST00000420034.1
ENST00000372842.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr17_-_7108436 | 1.36 |
ENST00000493294.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr5_+_152870734 | 1.36 |
ENST00000521843.2
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr3_+_120626919 | 1.36 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chr1_+_201708992 | 1.35 |
ENST00000367295.1
|
NAV1
|
neuron navigator 1 |
| chr2_+_177053307 | 1.35 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr15_+_68871569 | 1.35 |
ENST00000566799.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr19_+_50194821 | 1.35 |
ENST00000594587.1
ENST00000595969.1 |
CPT1C
|
carnitine palmitoyltransferase 1C |
| chr10_-_100995540 | 1.34 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chr3_-_73610759 | 1.34 |
ENST00000466780.1
|
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr8_-_57233103 | 1.34 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr12_+_20848486 | 1.33 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr2_-_68547061 | 1.33 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chrX_-_153095813 | 1.32 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr11_-_89224299 | 1.31 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr3_+_158991025 | 1.29 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr15_-_79383102 | 1.29 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr14_-_22005018 | 1.29 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr11_-_86383157 | 1.22 |
ENST00000393324.3
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr6_+_44238203 | 1.22 |
ENST00000451188.2
|
TMEM151B
|
transmembrane protein 151B |
| chr11_+_107461804 | 1.22 |
ENST00000531234.1
|
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr12_+_108523133 | 1.22 |
ENST00000547525.1
|
WSCD2
|
WSC domain containing 2 |
| chr16_+_72459838 | 1.19 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr7_-_150675372 | 1.18 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr5_+_125758813 | 1.18 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr4_-_99578776 | 1.16 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr16_+_76311169 | 1.16 |
ENST00000307431.8
ENST00000377504.4 |
CNTNAP4
|
contactin associated protein-like 4 |
| chr10_-_15413035 | 1.15 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chr15_+_71185148 | 1.15 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr14_-_105420241 | 1.14 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr17_-_1029866 | 1.14 |
ENST00000570525.1
|
ABR
|
active BCR-related |
| chr12_-_6809543 | 1.13 |
ENST00000540656.1
|
PIANP
|
PILR alpha associated neural protein |
| chr11_-_89224139 | 1.11 |
ENST00000413594.2
|
NOX4
|
NADPH oxidase 4 |
| chr4_-_13546632 | 1.11 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr17_+_26800648 | 1.11 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr1_-_168698433 | 1.11 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr5_+_125758865 | 1.10 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr19_+_40973049 | 1.09 |
ENST00000598249.1
ENST00000338932.3 ENST00000344104.3 |
SPTBN4
|
spectrin, beta, non-erythrocytic 4 |
| chr5_-_60140089 | 1.09 |
ENST00000507047.1
ENST00000438340.1 ENST00000425382.1 ENST00000508821.1 |
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr17_+_12569306 | 1.08 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr1_+_2398876 | 1.08 |
ENST00000449969.1
|
PLCH2
|
phospholipase C, eta 2 |
| chr2_-_68547019 | 1.08 |
ENST00000409862.1
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr22_+_38609538 | 1.07 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr8_-_140715294 | 1.07 |
ENST00000303015.1
ENST00000520439.1 |
KCNK9
|
potassium channel, subfamily K, member 9 |
| chr5_+_140201183 | 1.06 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr12_-_57443886 | 1.05 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr11_+_107461948 | 1.04 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chrX_-_153095945 | 1.03 |
ENST00000164640.4
|
PDZD4
|
PDZ domain containing 4 |
| chr4_-_99578789 | 1.03 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr5_+_56910048 | 1.01 |
ENST00000509844.1
|
CTD-2023N9.3
|
CTD-2023N9.3 |
| chr17_-_40333099 | 1.01 |
ENST00000607371.1
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr17_-_79139817 | 1.01 |
ENST00000326724.4
|
AATK
|
apoptosis-associated tyrosine kinase |
| chr1_-_193155729 | 1.00 |
ENST00000367434.4
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr6_-_55740352 | 1.00 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr3_-_112360116 | 0.99 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr14_-_22005197 | 0.99 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr17_+_26800756 | 0.98 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr3_-_98235962 | 0.98 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr3_-_64431058 | 0.98 |
ENST00000564377.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr19_+_19639670 | 0.98 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr2_+_115822233 | 0.97 |
ENST00000393146.2
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr12_+_6982725 | 0.97 |
ENST00000433346.1
|
LRRC23
|
leucine rich repeat containing 23 |
| chr17_-_7120525 | 0.97 |
ENST00000447163.1
ENST00000399506.2 ENST00000302955.6 |
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr2_-_73511559 | 0.96 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr7_+_23286182 | 0.96 |
ENST00000258733.4
ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB
|
glycoprotein (transmembrane) nmb |
| chr1_+_156611704 | 0.96 |
ENST00000329117.5
|
BCAN
|
brevican |
| chr11_+_19798964 | 0.96 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr6_-_131321863 | 0.95 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr11_-_89224488 | 0.95 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr14_+_85996507 | 0.94 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr4_-_114900126 | 0.94 |
ENST00000541197.1
|
ARSJ
|
arylsulfatase family, member J |
| chr8_-_23261589 | 0.93 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr12_+_31079362 | 0.93 |
ENST00000545802.1
|
TSPAN11
|
tetraspanin 11 |
| chr1_+_24646263 | 0.93 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr11_-_86383370 | 0.93 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr11_-_128894053 | 0.92 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_-_70992888 | 0.91 |
ENST00000320755.7
|
COL9A1
|
collagen, type IX, alpha 1 |
| chr8_-_57359131 | 0.90 |
ENST00000518974.1
ENST00000523051.1 ENST00000518770.1 ENST00000451791.2 |
PENK
|
proenkephalin |
| chr15_+_73976545 | 0.90 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr1_+_26485511 | 0.90 |
ENST00000374268.3
|
FAM110D
|
family with sequence similarity 110, member D |
| chr4_-_186732048 | 0.88 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_24645807 | 0.87 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr17_-_9862772 | 0.87 |
ENST00000580865.1
ENST00000583882.1 |
GAS7
|
growth arrest-specific 7 |
| chr15_+_54903673 | 0.86 |
ENST00000560537.1
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr14_-_88459503 | 0.86 |
ENST00000393568.4
ENST00000261304.2 |
GALC
|
galactosylceramidase |
| chr6_-_70992755 | 0.85 |
ENST00000370499.4
|
COL9A1
|
collagen, type IX, alpha 1 |
| chr1_+_24645865 | 0.85 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr5_-_147211226 | 0.85 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr11_-_89224508 | 0.85 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr17_-_41985096 | 0.84 |
ENST00000269095.4
ENST00000523220.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr5_+_176057663 | 0.83 |
ENST00000318682.6
|
EIF4E1B
|
eukaryotic translation initiation factor 4E family member 1B |
| chr6_+_39760783 | 0.83 |
ENST00000398904.2
ENST00000538976.1 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr1_+_39670360 | 0.82 |
ENST00000494012.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr11_-_89223883 | 0.82 |
ENST00000528341.1
|
NOX4
|
NADPH oxidase 4 |
| chr17_+_12569472 | 0.82 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr2_-_68546530 | 0.81 |
ENST00000409559.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr3_-_149051444 | 0.81 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr17_-_19290117 | 0.81 |
ENST00000497081.2
|
MFAP4
|
microfibrillar-associated protein 4 |
| chr19_-_9226439 | 0.80 |
ENST00000293614.1
ENST00000541538.1 |
OR7G1
|
olfactory receptor, family 7, subfamily G, member 1 |
| chr2_-_36779411 | 0.79 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr2_+_204193149 | 0.79 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr5_+_139505520 | 0.79 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr12_-_57081940 | 0.79 |
ENST00000436399.2
|
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr10_-_97200772 | 0.78 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr19_+_37997812 | 0.78 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr11_-_118966167 | 0.78 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr1_-_203320617 | 0.78 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr4_+_144312659 | 0.78 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr1_+_101185290 | 0.77 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr7_+_47834880 | 0.76 |
ENST00000258776.4
|
C7orf69
|
chromosome 7 open reading frame 69 |
| chr5_-_60139347 | 0.76 |
ENST00000511799.1
|
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr6_+_126102292 | 0.75 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_+_98264571 | 0.74 |
ENST00000607823.1
|
RP11-436D23.1
|
RP11-436D23.1 |
| chr14_+_85996471 | 0.74 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_24646002 | 0.73 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr17_-_1029052 | 0.73 |
ENST00000574437.1
|
ABR
|
active BCR-related |
| chr2_-_86564776 | 0.73 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr7_-_142583478 | 0.72 |
ENST00000436401.1
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr11_-_62752455 | 0.71 |
ENST00000360421.4
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr15_+_68871308 | 0.70 |
ENST00000261861.5
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr17_+_44076616 | 0.70 |
ENST00000537309.1
|
STH
|
saitohin |
| chr14_+_38033252 | 0.69 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr8_+_28351707 | 0.68 |
ENST00000537916.1
ENST00000523546.1 ENST00000240093.3 |
FZD3
|
frizzled family receptor 3 |
| chr2_-_190044480 | 0.68 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr15_+_25200074 | 0.67 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr1_+_24645915 | 0.67 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr11_-_62752429 | 0.67 |
ENST00000377871.3
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr18_+_32621324 | 0.67 |
ENST00000300249.5
ENST00000538170.2 ENST00000588910.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr16_-_75284758 | 0.67 |
ENST00000561970.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr3_+_113616317 | 0.67 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF652
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.6 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 1.8 | 5.5 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 1.0 | 3.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.9 | 3.7 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.9 | 6.5 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.9 | 2.6 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.8 | 2.3 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.7 | 2.0 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.5 | 2.6 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.5 | 1.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.5 | 1.9 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.4 | 1.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 3.5 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.4 | 1.4 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.3 | 0.7 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.3 | 1.0 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.3 | 6.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.3 | 3.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 2.9 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 1.7 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 2.5 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 1.8 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.2 | 1.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.2 | 0.9 | GO:0018277 | protein deamination(GO:0018277) |
| 0.2 | 2.3 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 3.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 0.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 4.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 4.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 11.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 0.9 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.2 | 2.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 4.1 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 0.8 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 2.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.9 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 3.8 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.9 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.1 | 1.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.7 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.1 | 1.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.8 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 1.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 2.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.6 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 1.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 1.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.3 | GO:1904238 | mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) pericyte cell differentiation(GO:1904238) |
| 0.1 | 1.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 2.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 1.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.9 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.1 | 3.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.4 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.7 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 4.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 1.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.4 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.4 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.1 | 3.4 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 0.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 1.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.4 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.3 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.1 | 0.4 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.6 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 1.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 1.4 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.1 | 0.8 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 1.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 1.2 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 1.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 4.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 1.0 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 1.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.1 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 1.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 1.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.3 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.3 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 1.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 1.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 1.1 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 1.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.3 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 1.8 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 1.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 2.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 1.8 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 1.5 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.5 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 1.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.8 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 2.9 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 2.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 1.0 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 1.0 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 1.4 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 2.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 0.0 | 0.6 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.5 | GO:0090662 | ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 2.2 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.8 | 2.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.5 | 2.9 | GO:0044308 | axonal spine(GO:0044308) |
| 0.3 | 1.8 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.3 | 6.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 0.8 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 4.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 2.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.2 | 0.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.3 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.2 | 9.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 2.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 0.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 7.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 2.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 4.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 5.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 1.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 3.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.8 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 1.7 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.5 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 4.9 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 1.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 19.2 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 3.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 6.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.1 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.9 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 10.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 1.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.6 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 2.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.8 | 5.5 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 1.2 | 3.6 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.9 | 3.8 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.7 | 2.8 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.6 | 2.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.5 | 6.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 1.4 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.5 | 2.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 3.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 6.0 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.4 | 3.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.4 | 2.5 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 3.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 1.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.3 | 1.0 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.3 | 3.9 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 6.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 4.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 2.9 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 2.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 1.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 3.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 1.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 1.9 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 1.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.2 | 0.7 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.2 | 1.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 0.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.5 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 3.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 2.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.6 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.1 | 1.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.6 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 3.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 6.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.9 | GO:0008430 | selenium binding(GO:0008430) phosphate ion binding(GO:0042301) |
| 0.1 | 0.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 2.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 1.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 2.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 1.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 2.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 3.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 4.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 1.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 6.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 1.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 1.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 2.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.1 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 2.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.8 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 8.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 6.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 11.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 3.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 4.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 9.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 4.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 2.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 2.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 2.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 3.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |