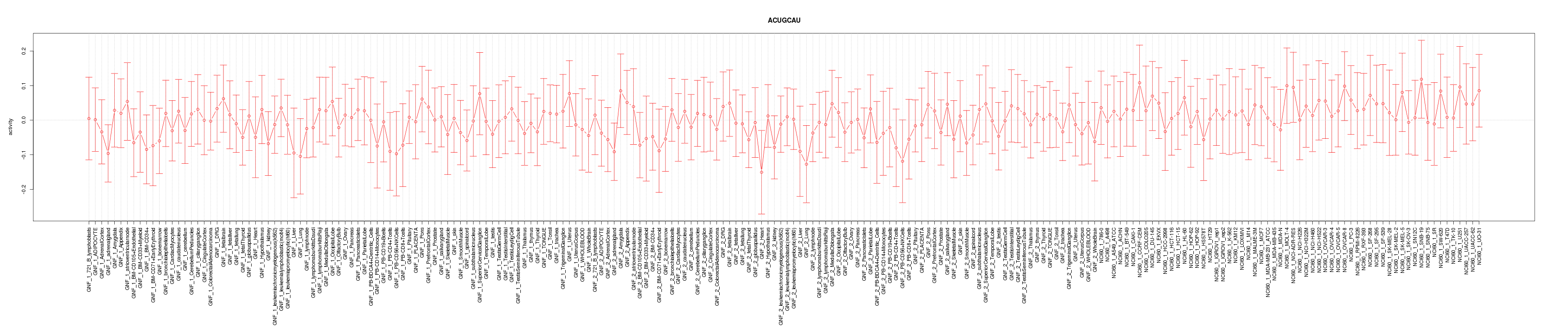
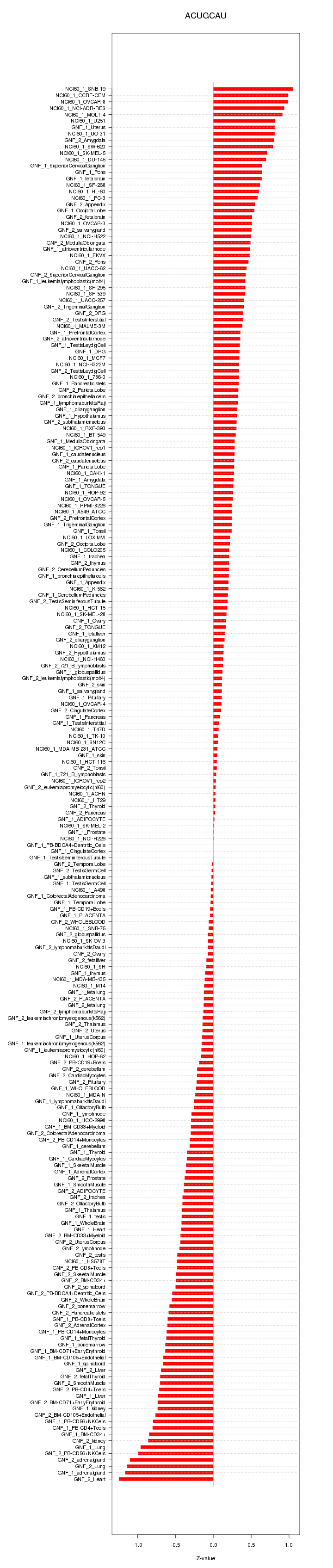
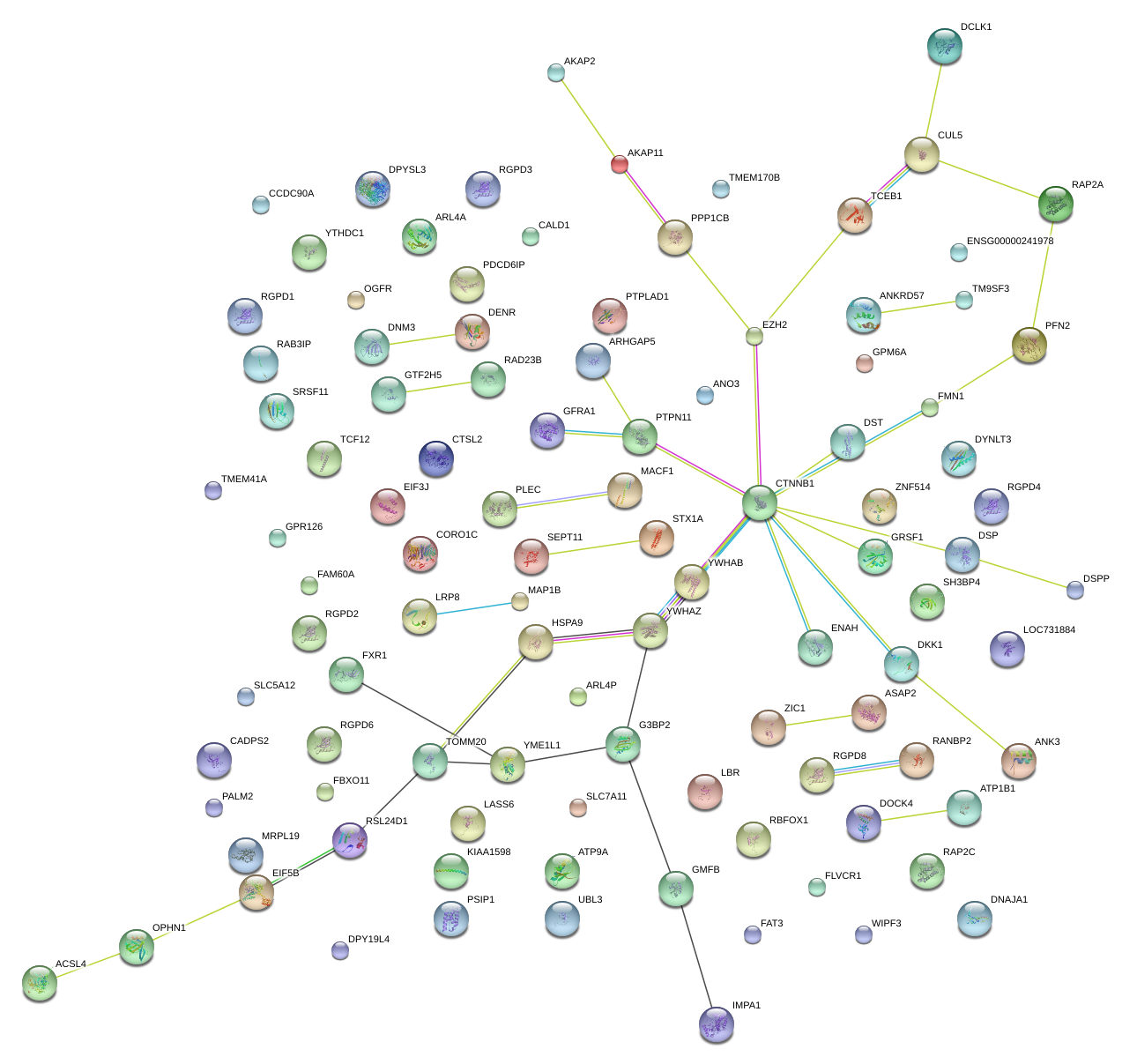
|
chr10_+_54074037
|
4.853
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr1_-_225840660
|
3.212
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr7_+_134464161
|
2.880
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr9_+_112887780
|
2.874
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr20_-_50384899
|
2.843
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr9_+_112810877
|
2.708
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr12_-_109125289
|
2.599
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr4_-_176923572
|
2.374
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr15_+_65822738
|
2.346
|
NM_016395
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1
|
|
chr4_+_77870864
|
2.278
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr2_+_235860616
|
2.050
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr3_-_149688638
|
2.044
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr9_+_110045516
|
2.013
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr12_-_31479120
|
1.974
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr3_+_180630054
|
1.926
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr5_+_71403040
|
1.847
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr14_-_54955714
|
1.819
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr15_+_57210823
|
1.781
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr9_+_112542496
|
1.739
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr2_+_9346876
|
1.733
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr12_+_70172748
|
1.658
|
NM_001024647
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr9_-_15510899
|
1.555
|
NM_001128217
NM_033222
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr4_-_176734181
|
1.542
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr20_+_43514340
|
1.517
|
NM_003404
NM_139323
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr12_+_70133179
|
1.482
|
NM_175623
NM_175625
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr3_+_147127151
|
1.400
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr15_+_57511616
|
1.373
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr1_+_169075869
|
1.317
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr5_-_137911317
|
1.311
|
NM_004134
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr12_+_70132562
|
1.265
|
NM_022456
NM_175624
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr11_+_26353677
|
1.237
|
NM_031418
|
ANO3
|
anoctamin 3
|
|
chr6_+_142622797
|
1.217
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr13_-_36705513
|
1.199
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr4_-_76598289
|
1.177
|
NM_012297
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr2_-_48132656
|
1.169
|
NM_001190274
|
FBXO11
|
F-box protein 11
|
|
chr16_+_6069094
|
1.112
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr4_-_139163222
|
1.107
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr10_-_98346793
|
1.099
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_+_70671331
|
1.055
|
NM_001190987
NM_004768
|
SRSF11
|
serine/arginine-rich splicing factor 11
|
|
chr2_+_28974608
|
1.052
|
NM_206876
NM_002709
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme
|
|
chr7_-_148580600
|
1.039
|
NM_001203249
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr3_-_185216545
|
1.032
|
NM_080652
|
TMEM41A
|
transmembrane protein 41A
|
|
chr2_+_109335906
|
1.022
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr10_-_118765087
|
1.020
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr9_+_33025207
|
1.020
|
NM_001539
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr8_-_74884142
|
1.018
|
NM_001204861
NM_001204862
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr11_+_92085261
|
1.003
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr1_+_213031596
|
1.000
|
NM_014053
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1
|
|
chr4_-_76598611
|
1.000
|
NM_203504
NM_203505
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr7_-_111846384
|
0.990
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr1_-_225616556
|
0.990
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr8_-_74884500
|
0.986
|
NM_001204857
NM_001204858
NM_001204859
NM_001204860
NM_001204863
NM_001204864
NM_005648
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr10_-_27443309
|
0.982
|
NM_001253866
NM_014263
NM_139312
|
YME1L1
|
YME1-like 1 (S. cerevisiae)
|
|
chr6_+_11538486
|
0.980
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr1_+_171810611
|
0.973
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr1_-_235292206
|
0.945
|
NM_014765
|
TOMM20
|
translocase of outer mitochondrial membrane 20 homolog (yeast)
|
|
chr15_-_55489128
|
0.934
|
NM_016304
|
RSL24D1
|
ribosomal L24 domain containing 1
|
|
chr7_+_134576150
|
0.928
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr2_+_75873907
|
0.926
|
NM_014763
|
MRPL19
|
mitochondrial ribosomal protein L19
|
|
chr11_+_107879407
|
0.911
|
NM_003478
|
CUL5
|
cullin 5
|
|
chr13_+_42846255
|
0.909
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr10_-_62493282
|
0.905
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr8_-_82598517
|
0.893
|
NM_001144878
NM_001144879
NM_005536
|
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1
|
|
chr3_+_41240922
|
0.882
|
NM_001098209
NM_001098210
NM_001904
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa
|
|
chrX_-_37706816
|
0.866
|
NM_006520
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr7_-_122526781
|
0.858
|
NM_001009571
NM_001167940
NM_017954
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chrX_-_108976509
|
0.858
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr2_+_169312793
|
0.846
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chrX_-_67653169
|
0.818
|
NM_002547
|
OPHN1
|
oligophrenin 1
|
|
chr10_-_118032717
|
0.818
|
NM_001145453
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr13_-_30424795
|
0.817
|
NM_007106
|
UBL3
|
ubiquitin-like 3
|
|
chr1_-_53793743
|
0.788
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr6_-_13814546
|
0.787
|
NM_001031713
|
CCDC90A
|
coiled-coil domain containing 90A
|
|
chr7_+_29846112
|
0.786
|
NM_001080529
|
WIPF3
|
WAS/WASL interacting protein family, member 3
|
|
chr4_-_71705188
|
0.762
|
NM_001098477
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr10_-_62332666
|
0.752
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr14_+_32546445
|
0.750
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr2_+_110371887
|
0.746
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chrX_-_131351917
|
0.740
|
NM_021183
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr7_+_12726910
|
0.737
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr4_-_69215699
|
0.727
|
NM_001031732
NM_133370
|
YTHDC1
|
YTH domain containing 1
|
|
chr15_+_44829265
|
0.722
|
NM_003758
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr6_+_158589378
|
0.717
|
NM_207118
|
GTF2H5
|
general transcription factor IIH, polypeptide 5
|
|
chr9_-_99801365
|
0.715
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr3_+_33840060
|
0.708
|
NM_001162429
NM_013374
|
PDCD6IP
|
programmed cell death 6 interacting protein
|
|
chr10_-_118031542
|
0.700
|
NM_145793
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr12_+_112856535
|
0.697
|
NM_002834
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr5_-_146833150
|
0.691
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr4_-_71705613
|
0.689
|
NM_002092
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr10_-_118032975
|
0.687
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr1_+_39547088
|
0.682
|
NM_012090
|
MACF1
|
microtubule-actin crosslinking factor 1
|
|
chr2_-_48115846
|
0.679
|
NM_025133
|
FBXO11
|
F-box protein 11
|
|
chr12_+_123237302
|
0.677
|
NM_003677
|
DENR
|
density-regulated protein
|
|
chr8_+_95732101
|
0.674
|
NM_181787
|
DPY19L4
|
dpy-19-like 4 (C. elegans)
|
|
chr7_+_12726413
|
0.673
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr2_-_95825262
|
0.670
|
NM_032788
|
ZNF514
|
zinc finger protein 514
|
|
chr11_-_26743545
|
0.667
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr9_-_99801924
|
0.667
|
NM_001201575
|
CTSL2
|
cathepsin L2
|
|
chr15_-_33360084
|
0.667
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr2_+_99953749
|
0.666
|
NM_015904
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr7_-_73133917
|
0.664
|
NM_001165903
NM_004603
|
STX1A
|
syntaxin 1A (brain)
|
|
chr6_+_7541863
|
0.657
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr20_+_19870209
|
0.651
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr16_-_3627353
|
0.626
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr17_-_36105005
|
0.621
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr1_+_224301790
|
0.616
|
NM_001136115
NM_015176
|
FBXO28
|
F-box protein 28
|
|
chr20_+_56884749
|
0.612
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr17_-_60142598
|
0.610
|
NM_005121
|
MED13
|
mediator complex subunit 13
|
|
chr20_+_19867075
|
0.590
|
NM_001242581
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_+_212208918
|
0.581
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr4_+_39699663
|
0.579
|
NM_001111112
NM_001111113
NM_005339
|
UBE2K
|
ubiquitin-conjugating enzyme E2K
|
|
chr6_+_163835668
|
0.575
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr1_-_225615787
|
0.574
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chr16_+_53088791
|
0.573
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr16_+_7382750
|
0.565
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr13_-_36429798
|
0.561
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr7_-_32529972
|
0.559
|
NM_012322
|
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr15_+_45879416
|
0.558
|
NM_012388
|
PLDN
|
pallidin homolog (mouse)
|
|
chr11_+_125462736
|
0.558
|
NM_152713
|
STT3A
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr7_-_93633527
|
0.557
|
NM_005868
|
BET1
|
blocked early in transport 1 homolog (S. cerevisiae)
|
|
chr2_-_203736223
|
0.554
|
NM_138468
|
ICA1L
|
islet cell autoantigen 1,69kDa-like
|
|
chr7_-_32534869
|
0.554
|
NM_001130710
NM_001139499
|
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr1_-_160492982
|
0.553
|
NM_001184714
NM_001184715
NM_001184716
NM_052931
|
SLAMF6
|
SLAM family member 6
|
|
chr5_+_179125796
|
0.552
|
NM_001024649
NM_001746
|
CANX
|
calnexin
|
|
chr16_+_67063035
|
0.529
|
NM_001755
NM_022845
|
CBFB
|
core-binding factor, beta subunit
|
|
chr16_+_12995454
|
0.527
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chrX_-_20284708
|
0.526
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr7_-_148581403
|
0.522
|
NM_001203247
NM_001203248
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr2_-_37544165
|
0.518
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr11_+_93517394
|
0.514
|
NM_004268
|
MED17
|
mediator complex subunit 17
|
|
chr2_+_118846029
|
0.513
|
NM_016133
|
INSIG2
|
insulin induced gene 2
|
|
chr8_+_104512975
|
0.506
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr14_+_89029690
|
0.502
|
NM_207661
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr4_-_100815666
|
0.501
|
NM_001243736
NM_021970
|
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3
|
|
chr8_-_56685869
|
0.499
|
NM_152417
|
TMEM68
|
transmembrane protein 68
|
|
chr5_+_149340287
|
0.496
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr6_+_64281910
|
0.493
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr2_+_70121074
|
0.489
|
NM_006857
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5)
|
|
chr3_-_160283361
|
0.485
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr11_-_85780899
|
0.484
|
NM_001206947
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr14_+_89060729
|
0.481
|
NM_207662
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr16_+_81069316
|
0.479
|
NM_015251
|
ATMIN
|
ATM interactor
|
|
chr3_+_173116243
|
0.478
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr13_+_27131799
|
0.475
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr12_-_63328663
|
0.472
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr2_+_149632772
|
0.471
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr6_-_7313352
|
0.469
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr12_-_25403824
|
0.462
|
NM_004985
NM_033360
|
KRAS
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
|
|
chr19_+_57078889
|
0.457
|
NM_001001668
|
ZNF470
|
zinc finger protein 470
|
|
chr14_+_89029252
|
0.455
|
NM_001160103
NM_001160104
NM_024824
NM_207660
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr6_-_80656870
|
0.448
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr1_+_218458628
|
0.444
|
NM_016052
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae)
|
|
chr5_-_146889618
|
0.434
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr14_-_95623665
|
0.431
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr3_+_141043054
|
0.429
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr3_+_36421944
|
0.421
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr15_+_50716568
|
0.407
|
NM_001128610
NM_001128611
NM_005154
|
USP8
|
ubiquitin specific peptidase 8
|
|
chr6_-_79787939
|
0.407
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr4_+_57775064
|
0.402
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr19_-_41255827
|
0.397
|
NM_198476
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr6_+_126240369
|
0.396
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr9_+_134305476
|
0.396
|
NM_013318
|
PRRC2B
|
proline-rich coiled-coil 2B
|
|
chr17_+_30469373
|
0.393
|
NM_001033566
NM_001033568
NM_018307
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr2_+_65215577
|
0.389
|
NM_001193493
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chrX_+_11129405
|
0.387
|
NM_001122608
NM_001171991
NM_005333
|
HCCS
|
holocytochrome c synthase
|
|
chrX_-_110039285
|
0.383
|
NM_001143981
NM_001143982
NM_001143983
|
CHRDL1
|
chordin-like 1
|
|
chr4_-_166034003
|
0.381
|
NM_001100389
|
TMEM192
|
transmembrane protein 192
|
|
chr17_-_30669133
|
0.380
|
NM_022344
|
C17orf75
|
chromosome 17 open reading frame 75
|
|
chr3_+_50712671
|
0.376
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr2_+_65216482
|
0.373
|
NM_003038
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr2_+_86333304
|
0.353
|
NM_017952
|
PTCD3
|
Pentatricopeptide repeat domain 3
|
|
chr11_-_85780105
|
0.343
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr7_+_117824085
|
0.343
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr14_-_95599794
|
0.342
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr4_+_57774035
|
0.338
|
NM_005612
|
REST
|
RE1-silencing transcription factor
|
|
chr1_+_65613771
|
0.335
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr15_-_101792125
|
0.335
|
NM_014918
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr16_+_6823809
|
0.334
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_+_167691156
|
0.326
|
NM_001146191
NM_003953
NM_024569
|
MPZL1
|
myelin protein zero-like 1
|
|
chr8_-_74659082
|
0.319
|
NM_001164380
NM_001164381
NM_001164382
NM_001164383
NM_001164385
NM_014393
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr6_-_149806116
|
0.315
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chrX_-_110038992
|
0.313
|
NM_145234
|
CHRDL1
|
chordin-like 1
|
|
chr8_+_99130067
|
0.305
|
NM_015029
|
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr10_+_124220990
|
0.305
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr1_-_24306832
|
0.304
|
NM_001191005
NM_001191006
NM_001191007
NM_001191009
NM_006625
NM_054016
|
SRSF10
|
serine/arginine-rich splicing factor 10
|
|
chr11_-_27528298
|
0.303
|
NM_018362
|
LIN7C
|
lin-7 homolog C (C. elegans)
|
|
chr5_-_79286793
|
0.301
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr4_-_186347037
|
0.294
|
NM_018359
|
UFSP2
|
UFM1-specific peptidase 2
|
|
chr5_-_133304274
|
0.287
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr12_+_120884178
|
0.285
|
NM_176818
|
GATC
|
glutamyl-tRNA(Gln) amidotransferase, subunit C homolog (bacterial)
|
|
chr1_+_201798281
|
0.278
|
NM_018085
|
IPO9
|
importin 9
|
|
chr7_+_130794854
|
0.277
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr18_+_56530060
|
0.276
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr1_-_212004113
|
0.276
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr11_+_118478305
|
0.275
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr1_+_65613211
|
0.269
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr3_+_179370779
|
0.261
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr14_+_45431333
|
0.258
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr11_+_59522531
|
0.256
|
NM_001178040
NM_004177
|
STX3
|
syntaxin 3
|
|
chr1_+_54519259
|
0.253
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|