|
chr6_+_34204647
|
5.945
|
NM_145901
NM_145902
|
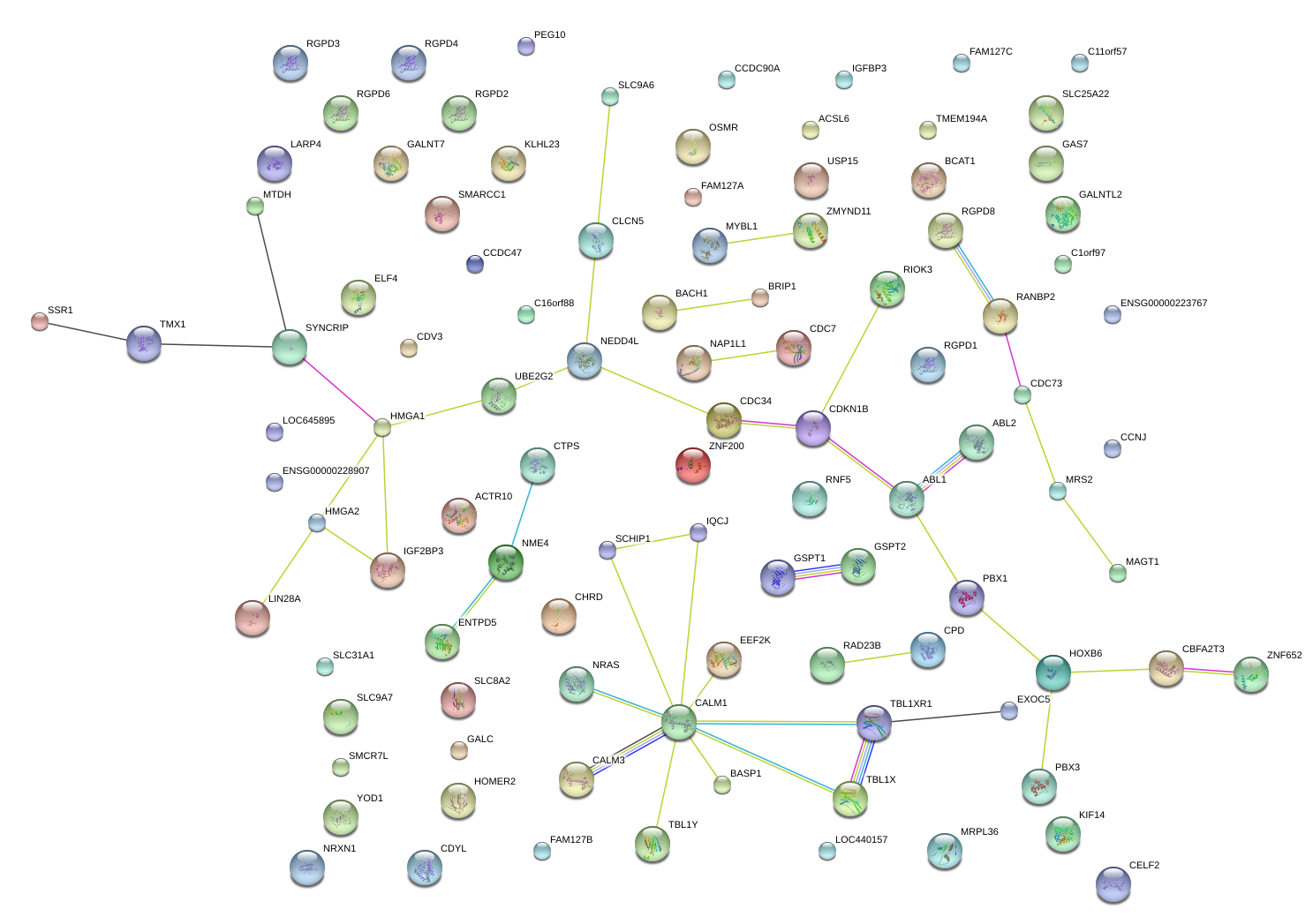
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_-_115259433
|
5.912
|
NM_002524
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr6_-_7313352
|
5.732
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr6_+_34204576
|
5.677
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_+_34206567
|
5.650
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr16_+_447175
|
4.862
|
NM_005009
|
NME4
|
non-metastatic cells 4, protein expressed in
|
|
chr3_+_133293256
|
4.613
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr9_+_128510477
|
4.466
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chrX_+_135067573
|
4.449
|
NM_001177651
NM_001042537
NM_006359
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr17_-_61850953
|
4.446
|
NM_020198
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr3_+_158991035
|
4.390
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr3_+_133292404
|
4.388
|
NM_001134422
NM_017548
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr7_+_94285620
|
4.367
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr7_+_94285676
|
4.314
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr6_-_86351156
|
4.108
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr12_-_76478737
|
4.063
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr6_-_86352635
|
3.854
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chrX_-_129244471
|
3.787
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr19_+_47104490
|
3.769
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr12_-_25055966
|
3.763
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr7_-_23509972
|
3.715
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr9_+_133589267
|
3.597
|
NM_007313
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr6_-_86352564
|
3.561
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_193091057
|
3.540
|
NM_024529
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr18_+_21032786
|
3.501
|
NM_003831
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr11_-_796175
|
3.499
|
NM_024698
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chrX_-_129244652
|
3.400
|
NM_001127197
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr12_-_25102281
|
3.386
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr18_+_55816546
|
3.299
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr12_-_25055228
|
3.260
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr1_+_41444968
|
3.189
|
NM_001905
|
CTPS
|
CTP synthase
|
|
chr1_-_207224298
|
3.155
|
NM_018566
|
YOD1
|
YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae)
|
|
chrX_-_134156488
|
3.124
|
NM_001078173
|
FAM127C
|
family with sequence similarity 127, member C
|
|
chr9_+_110045516
|
3.123
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr12_+_12869818
|
3.024
|
NM_004064
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr17_+_28707495
|
2.976
|
NM_001199775
|
CPD
|
carboxypeptidase D
|
|
chr9_+_115983807
|
2.937
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr22_+_39898248
|
2.911
|
NM_019008
|
SMCR7L
|
Smith-Magenis syndrome chromosome region, candidate 7-like
|
|
chr9_+_128509616
|
2.874
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr5_+_38845962
|
2.820
|
NM_003999
|
OSMR
|
oncostatin M receptor
|
|
chr10_+_97803064
|
2.778
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr12_+_66218141
|
2.762
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr3_+_159557649
|
2.761
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr6_-_86352982
|
2.675
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr18_+_55888750
|
2.633
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr16_-_89043215
|
2.614
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr3_-_47823386
|
2.612
|
NM_003074
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr21_+_30671217
|
2.544
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr12_+_62654120
|
2.510
|
NM_001252078
NM_001252079
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr4_+_174089889
|
2.509
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr14_-_88460008
|
2.485
|
NM_001201402
|
GALC
|
galactosylceramidase
|
|
chr14_+_51706885
|
2.450
|
NM_030755
|
TMX1
|
thioredoxin-related transmembrane protein 1
|
|
chr9_+_133710746
|
2.436
|
NM_005157
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr6_-_13814546
|
2.431
|
NM_001031713
|
CCDC90A
|
coiled-coil domain containing 90A
|
|
chr6_+_24403144
|
2.418
|
NM_020662
|
MRS2
|
MRS2 magnesium homeostasis factor homolog (S. cerevisiae)
|
|
chr18_+_55714457
|
2.388
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr14_+_58666818
|
2.290
|
NM_018477
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae)
|
|
chr12_+_50794591
|
2.245
|
NM_001170803
NM_001170804
NM_001170808
NM_052879
NM_199188
NM_199190
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr5_+_17217474
|
2.211
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr1_+_26737232
|
2.072
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr10_+_180404
|
2.030
|
NM_006624
NM_001202465
NM_001202466
NM_212479
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chrX_+_134166318
|
2.018
|
NM_001078171
|
FAM127A
|
family with sequence similarity 127, member A
|
|
chr14_+_90863302
|
2.012
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr21_-_30365111
|
1.981
|
NM_015565
|
LTN1
|
listerin E3 ubiquitin protein ligase 1
|
|
chr19_+_531688
|
1.945
|
NM_004359
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr2_+_109335906
|
1.906
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr3_+_184097860
|
1.902
|
NM_003741
|
CHRD
|
chordin
|
|
chr17_-_9929567
|
1.900
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr3_+_158787040
|
1.899
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr16_-_12010518
|
1.889
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr11_+_111944967
|
1.887
|
NM_001082969
NM_001082970
NM_018195
|
C11orf57
|
chromosome 11 open reading frame 57
|
|
chr12_-_57472573
|
1.872
|
NM_001130963
NM_015257
|
TMEM194A
|
transmembrane protein 194A
|
|
chr18_+_55862577
|
1.856
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_+_32146161
|
1.830
|
NM_006913
|
RNF5
|
ring finger protein 5
|
|
chr16_+_22217369
|
1.799
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr21_-_46221662
|
1.661
|
NM_001202489
NM_003343
NM_182688
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2
|
|
chr5_-_131347518
|
1.603
|
NM_001205248
NM_001205251
NM_001205250
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr11_-_798196
|
1.601
|
NM_001191061
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr18_+_55711388
|
1.551
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_+_4836331
|
1.441
|
NM_001143970
|
CDYL
|
chromodomain protein, Y-like
|
|
chr1_+_91966365
|
1.409
|
NM_001134419
NM_003503
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chr17_-_47439326
|
1.378
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr1_+_91966653
|
1.353
|
NM_001134420
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chr5_-_131347249
|
1.335
|
NM_001009185
NM_001205247
NM_015256
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr11_-_797955
|
1.297
|
NM_001191060
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr7_-_45960738
|
1.279
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr3_-_176915012
|
1.259
|
NM_024665
|
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1
|
|
chrX_-_77150907
|
1.247
|
NM_032121
|
MAGT1
|
magnesium transporter 1
|
|
chr8_+_98656329
|
1.229
|
NM_178812
|
MTDH
|
metadherin
|
|
chr16_-_12009804
|
1.227
|
NM_001130006
NM_002094
|
GSPT1
|
G1 to S phase transition 1
|
|
chr19_-_47975244
|
1.197
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr10_+_11206928
|
1.164
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr8_-_67525426
|
1.163
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr17_+_28705922
|
1.148
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr6_+_4890225
|
1.106
|
NM_001143971
|
CDYL
|
chromodomain protein, Y-like
|
|
chr14_-_57735614
|
1.076
|
NM_006544
|
EXOC5
|
exocyst complex component 5
|
|
chr10_+_181410
|
1.074
|
NM_001202464
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr2_+_170590241
|
1.064
|
NM_144711
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr2_-_50574891
|
1.033
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr1_-_200589856
|
1.028
|
NM_014875
|
KIF14
|
kinesin family member 14
|
|
chr17_-_46682333
|
0.995
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr16_-_3285359
|
0.915
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chrX_-_134185978
|
0.914
|
NM_001078172
NM_001134321
|
FAM127B
|
family with sequence similarity 127, member B
|
|
chrX_+_49832214
|
0.894
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr6_+_134274247
|
0.872
|
NM_004865
|
TBPL1
|
TBP-like 1
|
|
chr2_+_242641310
|
0.859
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr14_-_95599794
|
0.841
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr10_+_11059854
|
0.814
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_-_39664129
|
0.795
|
NM_003618
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr1_-_205601999
|
0.786
|
NM_001973
NM_021795
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr10_+_11047253
|
0.782
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr18_+_19749403
|
0.779
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr8_-_82754427
|
0.777
|
NM_022133
NM_152836
NM_152837
|
SNX16
|
sorting nexin 16
|
|
chr1_-_51984994
|
0.743
|
NM_001981
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr19_+_14063318
|
0.734
|
NM_138353
|
DCAF15
|
DDB1 and CUL4 associated factor 15
|
|
chr17_+_28268622
|
0.728
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr10_+_102672325
|
0.709
|
NM_001136123
NM_001243770
NM_018121
|
FAM178A
|
family with sequence similarity 178, member A
|
|
chr8_+_37594078
|
0.697
|
NM_001003791
NM_007175
|
ERLIN2
|
ER lipid raft associated 2
|
|
chr2_-_142889269
|
0.682
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chrX_+_67718623
|
0.677
|
NM_001195214
NM_173834
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr15_-_52861177
|
0.670
|
NM_006628
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr14_+_72398816
|
0.661
|
NM_001204423
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr2_-_144994905
|
0.659
|
NM_024659
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr2_-_145052059
|
0.641
|
NM_001006636
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr2_+_172378856
|
0.637
|
NM_001127383
NM_024843
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr3_+_101498285
|
0.635
|
NM_001134456
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr1_-_51887740
|
0.633
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr20_+_306205
|
0.619
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr2_-_51259536
|
0.619
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr3_+_101498028
|
0.611
|
NM_145037
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr14_-_68162419
|
0.607
|
NM_001252650
NM_016026
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis)
|
|
chr5_-_55290601
|
0.604
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr15_-_38856830
|
0.596
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr17_+_2699731
|
0.596
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr1_+_50569581
|
0.584
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_+_153232178
|
0.570
|
NM_000427
|
LOR
|
loricrin
|
|
chr1_+_50571963
|
0.569
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr12_-_106533810
|
0.562
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr18_-_24445652
|
0.561
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr7_+_7222142
|
0.549
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr11_+_10772781
|
0.527
|
NM_014633
|
CTR9
|
Ctr9, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr17_-_45266664
|
0.526
|
NM_001114091
NM_001256
|
CDC27
|
cell division cycle 27 homolog (S. cerevisiae)
|
|
chr3_+_50712671
|
0.520
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr1_-_53793743
|
0.513
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr15_+_52043738
|
0.499
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr17_-_48278987
|
0.494
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr13_-_102568994
|
0.483
|
NM_004115
|
FGF14
|
fibroblast growth factor 14
|
|
chr2_+_207630111
|
0.478
|
NM_001136193
NM_001136194
NM_014929
|
FASTKD2
|
FAST kinase domains 2
|
|
chr6_+_4776641
|
0.474
|
NM_004824
|
CDYL
|
chromodomain protein, Y-like
|
|
chr1_+_101003692
|
0.465
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr21_+_17102370
|
0.460
|
NM_013396
|
USP25
|
ubiquitin specific peptidase 25
|
|
chr21_+_30671736
|
0.457
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr18_+_9475528
|
0.451
|
NM_006788
|
RALBP1
|
ralA binding protein 1
|
|
chr1_+_179923870
|
0.449
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr12_-_69326946
|
0.446
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chr16_-_52581713
|
0.443
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr1_+_50574589
|
0.439
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr22_+_30279064
|
0.427
|
NM_021090
NM_153050
NM_153051
|
MTMR3
|
myotubularin related protein 3
|
|
chr9_-_77567735
|
0.426
|
NM_017998
|
C9orf40
|
chromosome 9 open reading frame 40
|
|
chr14_-_95623665
|
0.413
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr5_+_153825497
|
0.411
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr7_+_150068252
|
0.378
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr13_-_33859835
|
0.355
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_+_61547179
|
0.352
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr6_+_170102236
|
0.347
|
NM_001029863
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr16_+_31483066
|
0.345
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr4_-_90758126
|
0.327
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr3_-_53079285
|
0.322
|
NM_001005159
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr12_-_58240746
|
0.311
|
NM_005730
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr16_-_22385596
|
0.308
|
NM_001802
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa
|
|
chr17_-_37607314
|
0.298
|
NM_004774
|
MED1
|
mediator complex subunit 1
|
|
chr13_-_33760215
|
0.276
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_+_215178884
|
0.268
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr6_-_24911194
|
0.266
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr3_-_149688638
|
0.258
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr9_+_91003238
|
0.253
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr22_-_22221953
|
0.247
|
NM_002745
NM_138957
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr2_-_145090038
|
0.237
|
NM_001164629
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr4_-_52904424
|
0.235
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr4_-_90759446
|
0.233
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr12_+_123868608
|
0.222
|
NM_020382
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr1_-_103574051
|
0.217
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr8_+_59465727
|
0.203
|
NM_001007067
NM_001007068
NM_001007069
NM_001007070
NM_005625
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr14_+_72399149
|
0.201
|
NM_001204424
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr20_-_45035253
|
0.201
|
NM_133171
NM_182764
|
ELMO2
|
engulfment and cell motility 2
|
|
chr4_-_47840058
|
0.194
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr13_-_33780142
|
0.192
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr10_+_1102741
|
0.191
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr3_-_178789597
|
0.190
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr14_-_88459469
|
0.184
|
NM_000153
NM_001201401
|
GALC
|
galactosylceramidase
|
|
chrX_-_124097601
|
0.176
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1 (Drosophila)
|
|
chr13_-_103054027
|
0.172
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr17_-_60142598
|
0.168
|
NM_005121
|
MED13
|
mediator complex subunit 13
|
|
chr3_-_37034609
|
0.166
|
NM_014805
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1
|
|
chr1_+_99729847
|
0.166
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr8_-_28243941
|
0.165
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr14_-_95608054
|
0.164
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr16_+_70380763
|
0.157
|
NM_018332
|
DDX19A
|
DEAD (Asp-Glu-Ala-As) box polypeptide 19A
|
|
chr1_+_50513685
|
0.156
|
NM_001144777
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr8_+_22102618
|
0.148
|
NM_001722
|
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa
|