|
chr7_+_94024201
|
40.223
|
|
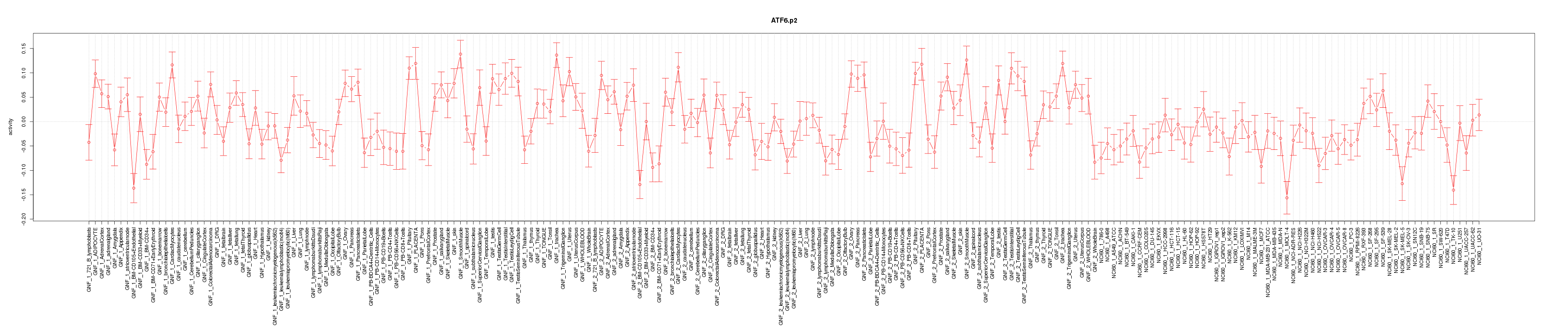
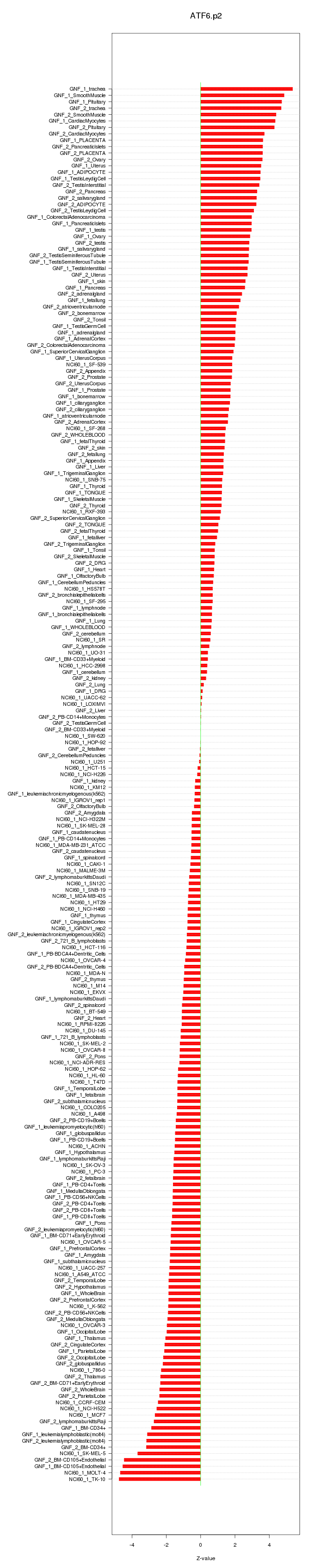
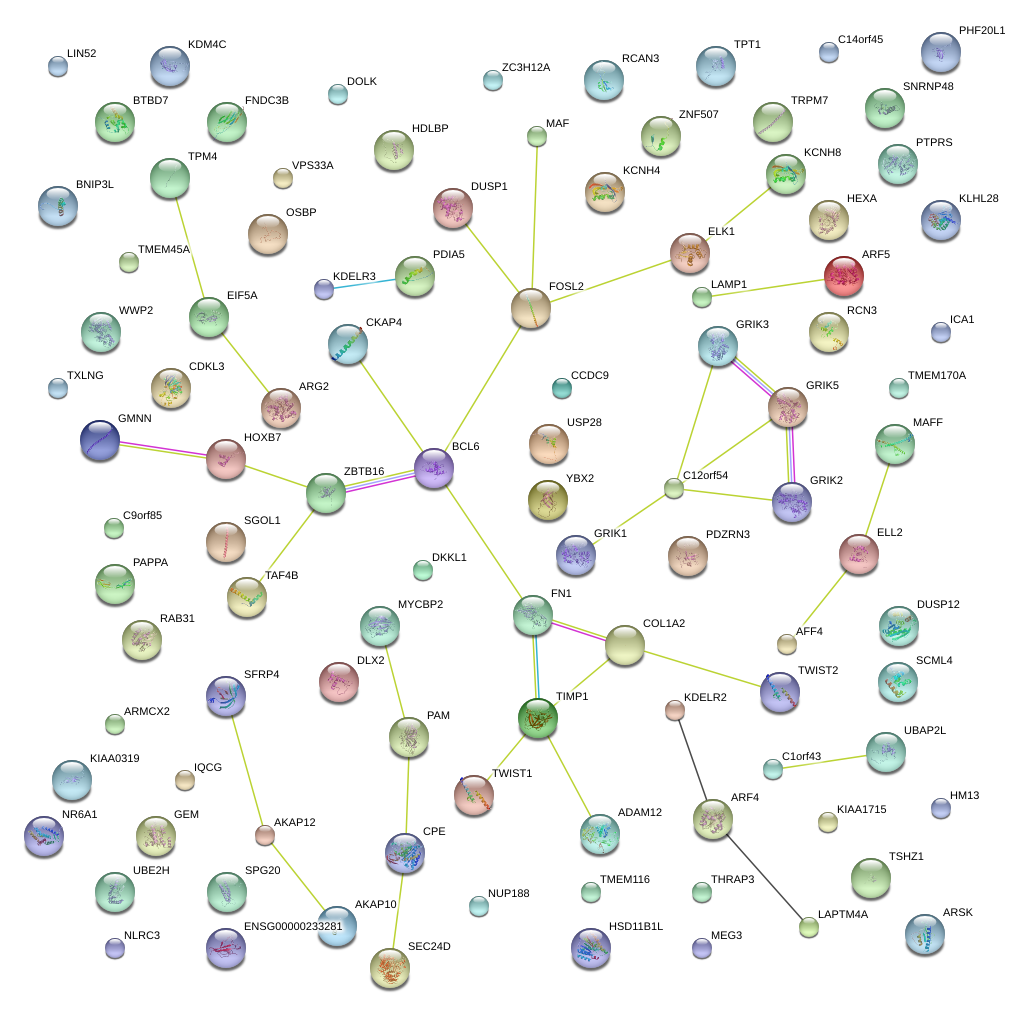
COL1A2
|
collagen, type I, alpha 2
|
|
chr7_+_94023872
|
28.277
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chrX_+_47441802
|
24.019
|
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr17_-_46688279
|
23.157
|
|
HOXB7
|
homeobox B7
|
|
chrX_+_47441650
|
21.055
|
NM_003254
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr7_+_64498643
|
20.937
|
|
CCT6P3
|
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 3
|
|
chr13_+_113951537
|
20.458
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr5_-_172198160
|
20.134
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr3_+_100211554
|
19.667
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr3_+_100211408
|
19.651
|
NM_018004
|
TMEM45A
|
transmembrane protein 45A
|
|
chr2_+_28615695
|
19.592
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr6_+_7590405
|
19.549
|
NM_152551
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12)
|
|
chr20_-_39766522
|
19.535
|
|
|
|
|
chr9_+_74526527
|
18.630
|
|
C9orf85
|
chromosome 9 open reading frame 85
|
|
chr6_-_108145507
|
18.009
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr7_-_6523689
|
17.887
|
NM_001100603
NM_006854
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr16_+_69796237
|
17.700
|
NM_007014
NM_199423
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr13_+_45915479
|
17.245
|
|
LOC100190939
|
uncharacterized LOC100190939
|
|
chr5_-_133706717
|
16.635
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr2_-_216300770
|
16.162
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr9_-_131709783
|
15.904
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr3_-_187463227
|
15.506
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_+_28615668
|
15.360
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr16_-_3627353
|
15.251
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr14_+_74551655
|
15.015
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr5_-_132299263
|
14.843
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chrX_-_100914816
|
14.840
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr18_+_72922725
|
14.510
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr2_+_28615771
|
14.467
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr3_-_187463474
|
14.097
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr11_-_59383179
|
13.852
|
|
OSBP
|
oxysterol binding protein
|
|
chr5_-_132299248
|
13.711
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr12_-_122751044
|
13.547
|
NM_022916
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr11_-_113746205
|
13.365
|
NM_020886
|
USP28
|
ubiquitin specific peptidase 28
|
|
chr3_-_57583153
|
13.316
|
NM_001660
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr12_-_122750954
|
12.986
|
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr13_+_113951589
|
12.932
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr2_-_172967627
|
12.753
|
|
DLX2
|
distal-less homeobox 2
|
|
chr19_+_50031232
|
12.284
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr17_-_7197896
|
12.073
|
|
YBX2
|
Y box binding protein 2
|
|
chr8_-_95274540
|
11.915
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr5_+_94890777
|
11.820
|
NM_198150
|
ARSK
|
arylsulfatase family, member K
|
|
chr8_-_95274520
|
11.731
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr17_-_7197811
|
11.699
|
|
YBX2
|
Y box binding protein 2
|
|
chr1_-_154193044
|
11.620
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr19_-_5340700
|
11.436
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr5_-_95297597
|
11.400
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr7_-_19157262
|
11.379
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr12_-_112450914
|
11.204
|
NM_001193453
NM_001193531
NM_138341
|
LOC728543
TMEM116
|
uncharacterized LOC728543
transmembrane protein 116
|
|
chr3_-_57583090
|
11.193
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr3_-_73673978
|
11.067
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr12_-_53448167
|
11.014
|
|
LOC283335
|
uncharacterized LOC283335
|
|
chr2_-_242254984
|
10.842
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr17_-_7197866
|
10.796
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr19_+_47759802
|
10.682
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr14_+_68086633
|
10.621
|
|
ARG2
|
arginase, type II
|
|
chr2_-_242255114
|
10.575
|
NM_005336
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr15_-_72668184
|
10.534
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr13_-_36920396
|
10.510
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr7_-_37956377
|
10.505
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr11_-_59383167
|
10.473
|
|
OSBP
|
oxysterol binding protein
|
|
chr20_+_30102261
|
10.404
|
|
HM13
|
histocompatibility (minor) 13
|
|
chr6_+_151561094
|
10.361
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr19_+_32836500
|
10.244
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr19_+_47759730
|
10.111
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr20_+_30102237
|
10.099
|
NM_030789
NM_178580
NM_178581
NM_178582
|
HM13
|
histocompatibility (minor) 13
|
|
chr7_-_129592794
|
10.098
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr3_-_197686837
|
9.935
|
NM_032263
|
IQCG
|
IQ motif containing G
|
|
chr5_+_102201713
|
9.913
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr3_+_171757432
|
9.756
|
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr8_+_26240522
|
9.756
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr9_-_127533397
|
9.471
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr2_-_216300444
|
9.364
|
|
FN1
|
fibronectin 1
|
|
chr9_+_131709970
|
9.237
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr19_+_16187266
|
9.230
|
|
TPM4
|
tropomyosin 4
|
|
chr12_-_106641675
|
9.230
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr6_+_30294224
|
9.205
|
|
|
|
|
chr1_+_113498984
|
9.160
|
|
LOC100506392
|
uncharacterized LOC100506392
|
|
chr9_+_6757636
|
8.889
|
NM_001146694
NM_001146695
NM_015061
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr4_-_119757183
|
8.860
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr7_-_37956515
|
8.838
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr3_-_8543327
|
8.831
|
|
LOC100288428
|
uncharacterized LOC100288428
|
|
chr19_+_49866986
|
8.824
|
NM_001197302
NM_001197301
NM_014419
|
DKKL1
|
dickkopf-like 1
|
|
chr11_-_59383616
|
8.764
|
NM_002556
|
OSBP
|
oxysterol binding protein
|
|
chr7_-_129592711
|
8.731
|
NM_001202498
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr12_+_112451229
|
8.723
|
|
|
|
|
chr3_-_57582866
|
8.704
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr13_-_45915277
|
8.686
|
NM_003295
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr14_-_45431121
|
8.659
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr9_+_131709991
|
8.626
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr7_-_8301767
|
8.465
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr16_-_79634610
|
8.452
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr4_+_166299942
|
8.396
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chrX_+_16804531
|
8.352
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr22_+_38597938
|
8.325
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr15_-_50978983
|
8.302
|
NM_017672
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr8_+_133787556
|
8.290
|
NM_016018
NM_032205
NM_198513
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr3_-_20227618
|
8.208
|
NM_001012409
NM_001012410
NM_001012411
NM_001012412
NM_001012413
NM_138484
NM_001199251
NM_001199252
NM_001199253
NM_001199254
NM_001199255
NM_001199256
NM_001199257
|
SGOL1
|
shugoshin-like 1 (S. pombe)
|
|
chr13_-_45915308
|
8.181
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr6_+_101846668
|
8.145
|
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr3_-_57583080
|
8.123
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr1_+_154193294
|
8.122
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr1_-_47184790
|
8.091
|
|
|
|
|
chr1_+_36689996
|
8.016
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr13_+_113951461
|
8.003
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr1_+_37940074
|
7.942
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr18_+_9708269
|
7.939
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr2_-_176866925
|
7.923
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chr19_+_5681016
|
7.914
|
NM_198704
NM_198705
NM_198533
NM_198707
NM_198706
NM_198708
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr11_+_113930430
|
7.904
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr18_+_23806385
|
7.850
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr14_+_101292451
|
7.812
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr17_+_7211260
|
7.812
|
NM_001143761
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr3_+_122785855
|
7.792
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr14_+_68086570
|
7.788
|
NM_001172
|
ARG2
|
arginase, type II
|
|
chr10_-_128077062
|
7.773
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr11_+_113930296
|
7.744
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr4_-_119757288
|
7.743
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr2_-_20251788
|
7.731
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr22_+_38864015
|
7.714
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr14_-_93799360
|
7.712
|
|
BTBD7
|
BTB (POZ) domain containing 7
|
|
chr16_-_75498582
|
7.598
|
NM_145254
|
TMEM170A
|
transmembrane protein 170A
|
|
chr17_-_19880991
|
7.567
|
NM_007202
|
AKAP10
|
A kinase (PRKA) anchor protein 10
|
|
chr14_+_74486049
|
7.564
|
NM_025057
|
C14orf45
|
chromosome 14 open reading frame 45
|
|
chrX_-_47509993
|
7.511
|
NM_001114123
NM_005229
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr16_+_68057144
|
7.507
|
NM_017803
|
DUS2L
|
dihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae)
|
|
chr3_-_119182423
|
7.440
|
NM_018266
|
TMEM39A
|
transmembrane protein 39A
|
|
chr8_+_104427590
|
7.413
|
|
DCAF13
|
DDB1 and CUL4 associated factor 13
|
|
chr6_+_127587984
|
7.398
|
|
RNF146
|
ring finger protein 146
|
|
chr10_-_101769640
|
7.397
|
NM_015221
|
DNMBP
|
dynamin binding protein
|
|
chr16_-_31470582
|
7.366
|
|
|
|
|
chr5_+_68389775
|
7.365
|
NM_001251969
NM_022902
NM_024055
|
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5
|
|
chr2_+_79347610
|
7.360
|
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr21_+_47545896
|
7.357
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr9_-_130477918
|
7.346
|
NM_001002913
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae)
|
|
chr7_-_8301681
|
7.295
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr2_-_220408264
|
7.288
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr1_+_160313172
|
7.285
|
|
NCSTN
|
nicastrin
|
|
chr19_+_16187315
|
7.282
|
|
TPM4
|
tropomyosin 4
|
|
chr17_+_7210929
|
7.280
|
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chrX_-_47509864
|
7.253
|
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr17_+_79885567
|
7.249
|
|
LOC92659
|
uncharacterized LOC92659
|
|
chr3_+_129159089
|
7.237
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr10_+_35416384
|
7.237
|
NM_181571
NM_183060
|
CREM
|
cAMP responsive element modulator
|
|
chr12_-_14720790
|
7.230
|
NM_024829
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr4_-_186456580
|
7.230
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr18_+_9708299
|
7.215
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr14_-_81687265
|
7.206
|
NM_015859
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa
|
|
chr13_-_45915203
|
7.163
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr8_+_128806720
|
7.131
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr3_+_129159138
|
7.113
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr3_+_129159150
|
7.087
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr5_+_102201526
|
7.079
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr10_-_128077011
|
7.037
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr1_+_160313061
|
7.009
|
NM_015331
|
NCSTN
|
nicastrin
|
|
chr7_-_73184535
|
6.975
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr6_+_31982538
|
6.899
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr11_-_35547144
|
6.873
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr9_+_6757919
|
6.866
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr3_+_129159075
|
6.863
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chrX_+_100805415
|
6.818
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr3_-_139108486
|
6.779
|
|
|
|
|
chr10_+_22634373
|
6.756
|
NM_001253854
NM_001253855
NM_012443
NM_172242
|
SPAG6
|
sperm associated antigen 6
|
|
chr12_+_28343390
|
6.756
|
|
|
|
|
chr1_+_36690038
|
6.731
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr3_+_129158949
|
6.721
|
NM_018262
NM_052985
NM_052989
NM_052990
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr7_+_116593354
|
6.701
|
NM_018412
NM_021908
|
ST7
|
suppression of tumorigenicity 7
|
|
chr6_-_70506472
|
6.680
|
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr7_+_128470462
|
6.671
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr19_+_18794487
|
6.656
|
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr14_+_101292483
|
6.630
|
|
|
|
|
chr4_-_54930706
|
6.629
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr12_+_56552141
|
6.606
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr13_-_95364248
|
6.572
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr7_+_99775346
|
6.566
|
|
STAG3
|
stromal antigen 3
|
|
chr17_-_13505217
|
6.562
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr10_+_121411071
|
6.554
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr11_-_119234859
|
6.551
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr10_+_121410751
|
6.516
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr16_-_1525006
|
6.505
|
|
CLCN7
|
chloride channel 7
|
|
chr1_-_35497249
|
6.493
|
|
ZMYM6
|
zinc finger, MYM-type 6
|
|
chr1_-_31712733
|
6.460
|
NM_024522
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1
|
|
chr12_-_14720553
|
6.450
|
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr2_-_211035913
|
6.409
|
NM_152519
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr14_-_64970319
|
6.396
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr10_+_121411042
|
6.388
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr10_+_121410891
|
6.386
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr1_-_235812994
|
6.363
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr2_-_1748285
|
6.343
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr8_+_104427637
|
6.324
|
|
DCAF13
|
DDB1 and CUL4 associated factor 13
|
|
chr7_+_44646180
|
6.299
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr17_-_46687925
|
6.295
|
|
HOXB7
|
homeobox B7
|
|
chr11_-_62598552
|
6.273
|
|
STX5
|
syntaxin 5
|
|
chr2_+_236402948
|
6.263
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr3_+_142442855
|
6.245
|
|
|
|
|
chr1_+_37940129
|
6.232
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr14_-_74485813
|
6.198
|
NM_001249
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5
|
|
chr7_-_72992888
|
6.193
|
|
TBL2
|
transducin (beta)-like 2
|
|
chr14_+_23305913
|
6.146
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr19_-_4457789
|
6.137
|
NM_025241
|
UBXN6
|
UBX domain protein 6
|