|
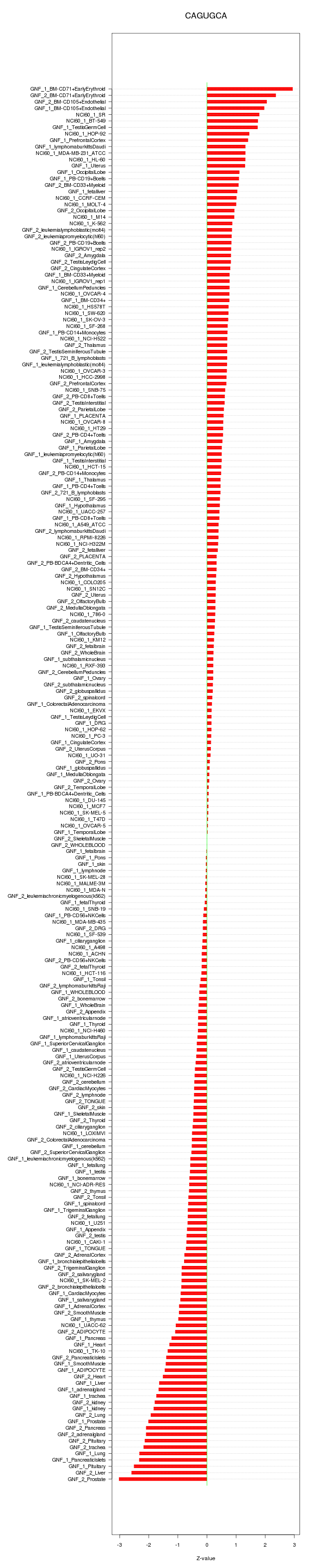
chr3_-_195808958
|
16.475
|
NM_003234
|
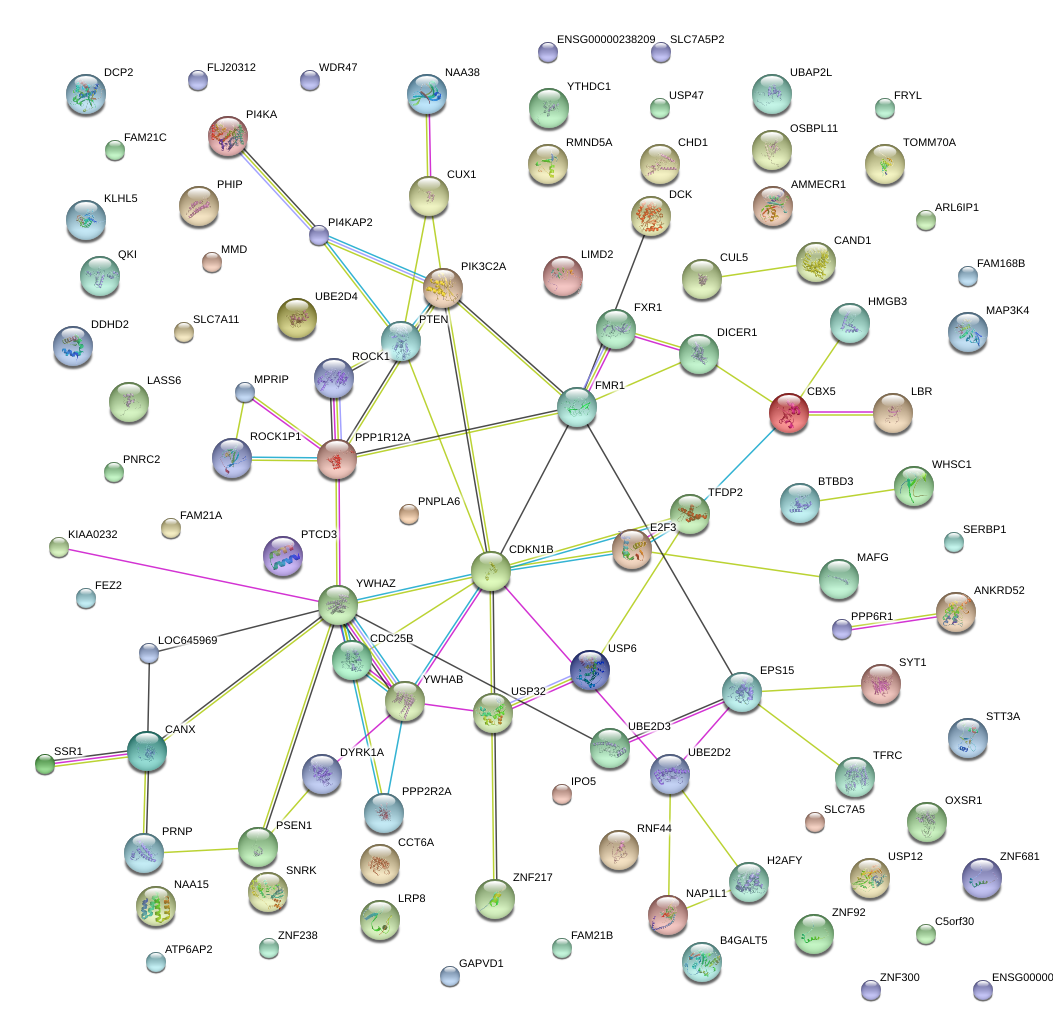
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr3_-_195808997
|
16.423
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr16_-_87903028
|
13.264
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr20_+_11898536
|
10.744
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr16_-_18812691
|
10.672
|
NM_015161
|
ARL6IP1
|
ADP-ribosylation factor-like 6 interacting protein 1
|
|
chr3_+_180630054
|
10.323
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chrX_-_109683457
|
9.855
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr7_+_56119377
|
9.774
|
NM_001009186
NM_001762
|
CCT6A
|
chaperonin containing TCP1, subunit 6A (zeta 1)
|
|
chr17_-_53499055
|
9.278
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr5_+_112312392
|
9.275
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr11_+_107879407
|
8.977
|
NM_003478
|
CUL5
|
cullin 5
|
|
chr20_+_43514340
|
8.921
|
NM_003404
NM_139323
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr5_-_134735565
|
8.141
|
NM_004893
|
H2AFY
|
H2A histone family, member Y
|
|
chr20_+_3776385
|
8.064
|
NM_004358
NM_021872
NM_021873
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr4_+_6784453
|
7.641
|
NM_001100590
NM_014743
|
KIAA0232
|
KIAA0232
|
|
chr2_+_169312793
|
7.378
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chr3_-_141719188
|
7.155
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chrX_-_109561314
|
7.087
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chrX_+_150151757
|
7.009
|
NM_005342
|
HMGB3
|
high mobility group box 3
|
|
chr3_-_141747434
|
6.989
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr6_+_163835668
|
6.916
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr3_+_38206968
|
6.650
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr1_-_225616556
|
6.594
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr11_+_11862955
|
6.387
|
NM_017944
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr13_+_98605928
|
6.260
|
NM_002271
|
IPO5
|
importin 5
|
|
chr1_-_225615787
|
6.077
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chr17_+_5031686
|
6.074
|
NM_004505
|
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene)
|
|
chr2_+_86333304
|
5.821
|
NM_017952
|
PTCD3
|
Pentatricopeptide repeat domain 3
|
|
chrX_+_40440145
|
5.713
|
NM_005765
|
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2
|
|
chr4_+_140222668
|
5.710
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr7_+_101460881
|
5.673
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr14_-_95599794
|
5.625
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr19_+_7599590
|
5.619
|
NM_001166111
NM_001166113
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr4_-_103748254
|
5.391
|
NM_181892
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr4_+_1894508
|
5.291
|
NM_007331
NM_133330
NM_133331
NM_133335
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr4_-_103748966
|
5.247
|
NM_003340
NM_181886
NM_181888
NM_181889
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr2_+_86947387
|
5.244
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr20_+_11871364
|
5.233
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr17_-_79881304
|
5.204
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr4_+_71859262
|
5.192
|
NM_000788
|
DCK
|
deoxycytidine kinase
|
|
chr2_-_131850988
|
5.179
|
NM_001009993
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr1_-_51984994
|
5.158
|
NM_001981
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr4_-_139163222
|
5.098
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr22_-_21088954
|
5.096
|
NM_002650
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr4_-_103748649
|
5.089
|
NM_181887
NM_181891
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr11_-_17191287
|
5.083
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr1_-_51887740
|
4.945
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr19_+_7599037
|
4.875
|
NM_006702
NM_001166112
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr1_-_67896098
|
4.863
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr12_-_54653369
|
4.776
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr6_-_79787939
|
4.772
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr12_+_67663060
|
4.599
|
NM_018448
|
CAND1
|
cullin-associated and neddylation-dissociated 1
|
|
chr12_+_12869818
|
4.571
|
NM_004064
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr3_-_125314367
|
4.571
|
NM_022776
|
OSBPL11
|
oxysterol binding protein-like 11
|
|
chr12_-_54653317
|
4.553
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr4_+_39063851
|
4.543
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chrX_+_146993450
|
4.523
|
NM_001185075
NM_001185076
NM_001185081
NM_001185082
NM_002024
|
FMR1
|
fragile X mental retardation 1
|
|
chr5_-_98262217
|
4.510
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr1_-_53793743
|
4.495
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr14_+_73603140
|
4.483
|
NM_000021
NM_007318
|
PSEN1
|
presenilin 1
|
|
chr12_-_76478737
|
4.423
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr3_-_141868208
|
4.273
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr20_-_52199635
|
4.260
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr20_-_48330417
|
4.228
|
NM_004776
|
B4GALT5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5
|
|
chr10_+_89623091
|
4.086
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr12_-_56652118
|
4.051
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr3_-_100120224
|
3.905
|
NM_014820
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae)
|
|
chr4_+_39046198
|
3.875
|
NM_001007075
NM_001171654
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr17_+_16945766
|
3.799
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr5_-_134734915
|
3.595
|
NM_001040158
NM_138609
NM_138610
|
H2AFY
|
H2A histone family, member Y
|
|
chr1_+_154192654
|
3.586
|
NM_001127320
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr4_-_48782281
|
3.542
|
NM_015030
|
FRYL
|
FRY-like
|
|
chr8_+_38089165
|
3.494
|
NM_001164232
|
DDHD2
|
DDHD domain containing 2
|
|
chr18_-_18691765
|
3.493
|
NM_005406
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr5_+_102594406
|
3.462
|
NM_033211
|
C5orf30
|
chromosome 5 open reading frame 30
|
|
chr5_+_179125796
|
3.440
|
NM_001024649
NM_001746
|
CANX
|
calnexin
|
|
chr21_+_38791206
|
3.418
|
NM_130436
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr8_+_26150731
|
3.409
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr1_+_24286300
|
3.405
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr14_-_95623665
|
3.380
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr12_-_80307690
|
3.378
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr20_+_4666796
|
3.375
|
NM_000311
NM_001080121
NM_001080122
NM_183079
NM_001080123
|
PRNP
|
prion protein
|
|
chr12_-_80329234
|
3.365
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr3_+_43328001
|
3.343
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr3_+_150321065
|
3.278
|
NM_016275
|
SELT
|
selenoprotein T
|
|
chr19_+_7600581
|
3.261
|
NM_001166114
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr9_+_128024109
|
3.223
|
NM_015635
|
GAPVD1
|
GTPase activating protein and VPS9 domains 1
|
|
chr1_-_109584589
|
3.218
|
NM_001142550
NM_001142551
NM_014969
|
WDR47
|
WD repeat domain 47
|
|
chr4_-_69215699
|
3.214
|
NM_001031732
NM_133370
|
YTHDC1
|
YTH domain containing 1
|
|
chr13_-_27745952
|
3.210
|
NM_182488
|
USP12
|
ubiquitin specific peptidase 12
|
|
chr19_-_55769920
|
3.196
|
NM_014931
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1
|
|
chr7_+_64838732
|
3.192
|
NM_007139
NM_152626
|
ZNF92
|
zinc finger protein 92
|
|
chr11_+_125462736
|
3.155
|
NM_152713
|
STT3A
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr6_+_161412807
|
3.154
|
NM_005922
NM_006724
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr6_+_20402040
|
3.144
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr5_-_175964223
|
3.136
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr7_+_117824085
|
3.124
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr2_-_36825234
|
3.102
|
NM_001042548
NM_005102
|
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II)
|
|
chr17_-_61777478
|
3.057
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr12_+_79258448
|
2.978
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr10_+_46222647
|
2.956
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
|
family with sequence similarity 21, member C
|
|
chr6_-_7313352
|
2.934
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr8_+_26149004
|
2.920
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr19_-_23941571
|
2.901
|
NM_138286
|
ZNF681
|
zinc finger protein 681
|
|
chr1_+_244216506
|
2.888
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr5_-_150284503
|
2.876
|
NM_001172831
NM_001172832
NM_052860
|
ZNF300
|
zinc finger protein 300
|
|
chr3_-_197463713
|
2.851
|
NM_014687
|
KIAA0226
|
KIAA0226
|
|
chr1_+_244214552
|
2.826
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr12_+_79439432
|
2.800
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr18_-_21017816
|
2.794
|
NM_032933
|
TMEM241
|
transmembrane protein 241
|
|
chr3_-_53079285
|
2.779
|
NM_001005159
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr5_+_72143929
|
2.776
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr12_+_77157842
|
2.741
|
NM_015336
|
ZDHHC17
|
zinc finger, DHHC-type containing 17
|
|
chr6_+_15246299
|
2.710
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr1_-_115259433
|
2.684
|
NM_002524
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr14_-_95608054
|
2.673
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chrX_+_103031753
|
2.663
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr19_+_33864574
|
2.658
|
NM_001806
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma
|
|
chr12_+_79257772
|
2.658
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr9_-_8733893
|
2.648
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chrX_+_103031433
|
2.634
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr4_-_103790002
|
2.599
|
NM_181893
NM_181890
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr2_+_231577536
|
2.582
|
NM_016289
|
CAB39
|
calcium binding protein 39
|
|
chr1_-_78225319
|
2.565
|
NM_015017
NM_201624
NM_201626
|
USP33
|
ubiquitin specific peptidase 33
|
|
chr6_-_86352635
|
2.549
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_116184573
|
2.547
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr2_+_33359607
|
2.542
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr5_+_52856460
|
2.521
|
NM_002495
|
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase)
|
|
chr2_+_231578262
|
2.506
|
NM_001130849
NM_001130850
|
CAB39
|
calcium binding protein 39
|
|
chr2_-_48132656
|
2.490
|
NM_001190274
|
FBXO11
|
F-box protein 11
|
|
chr12_-_56694174
|
2.479
|
NM_004077
|
CS
|
citrate synthase
|
|
chr4_+_1873035
|
2.444
|
NM_001042424
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr5_-_126366439
|
2.402
|
NM_178450
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chrX_-_134049248
|
2.398
|
NM_019556
|
MOSPD1
|
motile sperm domain containing 1
|
|
chr17_+_28707495
|
2.381
|
NM_001199775
|
CPD
|
carboxypeptidase D
|
|
chr6_+_64356430
|
2.379
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr6_-_86351156
|
2.368
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr14_+_23775970
|
2.363
|
NM_001199864
NM_004050
NM_001199839
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough
BCL2-like 2
|
|
chr3_+_5163929
|
2.335
|
NM_018184
|
ARL8B
|
ADP-ribosylation factor-like 8B
|
|
chr7_-_107204705
|
2.322
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr17_+_79373539
|
2.312
|
NM_001080519
|
BAHCC1
|
BAH domain and coiled-coil containing 1
|
|
chr1_-_156722023
|
2.311
|
NM_001126051
NM_001126050
|
HDGF
|
hepatoma-derived growth factor
|
|
chr12_-_46663207
|
2.306
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr5_+_72112417
|
2.296
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr5_-_133512641
|
2.294
|
NM_006930
NM_170679
|
SKP1
|
S-phase kinase-associated protein 1
|
|
chr7_+_50344255
|
2.280
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr19_-_10305645
|
2.272
|
NM_001130823
NM_001379
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1
|
|
chr15_-_65810020
|
2.263
|
NM_017743
NM_130434
NM_197961
|
DPP8
|
dipeptidyl-peptidase 8
|
|
chr14_-_55878537
|
2.251
|
NM_014924
|
ATG14
|
ATG14 autophagy related 14 homolog (S. cerevisiae)
|
|
chr12_-_46384365
|
2.242
|
NM_004719
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr2_-_64881040
|
2.221
|
NM_014755
|
SERTAD2
|
SERTA domain containing 2
|
|
chr1_+_116185249
|
2.200
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr7_-_104909442
|
2.194
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr6_-_86352564
|
2.187
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_26737232
|
2.181
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr5_+_96038492
|
2.163
|
NM_173060
|
CAST
|
calpastatin
|
|
chr22_-_21213099
|
2.138
|
NM_058004
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr15_-_65809603
|
2.134
|
NM_197960
|
DPP8
|
dipeptidyl-peptidase 8
|
|
chr12_-_57082056
|
2.131
|
NM_006601
|
PTGES3
|
prostaglandin E synthase 3 (cytosolic)
|
|
chr1_+_203595897
|
2.120
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr1_+_214776494
|
2.103
|
NM_016343
|
CENPF
|
centromere protein F, 350/400kDa (mitosin)
|
|
chr20_+_62887046
|
2.100
|
NM_001104925
NM_018257
|
PCMTD2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2
|
|
chr1_+_70671331
|
2.100
|
NM_001190987
NM_004768
|
SRSF11
|
serine/arginine-rich splicing factor 11
|
|
chr5_+_95997871
|
2.095
|
NM_001042440
|
CAST
|
calpastatin
|
|
chr1_+_12227050
|
2.068
|
NM_001066
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr7_-_1595746
|
2.068
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr9_-_36400212
|
2.055
|
NM_022781
NM_194329
|
RNF38
|
ring finger protein 38
|
|
chr1_+_118148506
|
2.036
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr4_-_24586178
|
2.007
|
NM_001358
|
DHX15
|
DEAH (Asp-Glu-Ala-His) box polypeptide 15
|
|
chr19_+_925984
|
1.995
|
NM_005224
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr4_+_128703402
|
1.985
|
NM_014278
|
HSPA4L
|
heat shock 70kDa protein 4-like
|
|
chrX_+_135067573
|
1.972
|
NM_001177651
NM_001042537
NM_006359
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr1_+_84944919
|
1.961
|
NM_025065
|
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae)
|
|
chr3_-_197476567
|
1.954
|
NM_001145642
|
KIAA0226
|
KIAA0226
|
|
chr2_-_132121730
|
1.947
|
NM_001077637
|
WTH3DI
|
RAB6C-like
|
|
chr3_+_115342150
|
1.945
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr15_+_91643181
|
1.926
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr14_+_96001297
|
1.921
|
NM_016417
|
GLRX5
|
glutaredoxin 5
|
|
chr13_-_79233266
|
1.919
|
NM_024546
|
RNF219
|
ring finger protein 219
|
|
chr13_-_99174251
|
1.910
|
NM_003576
|
STK24
|
serine/threonine kinase 24
|
|
chr2_-_48115846
|
1.899
|
NM_025133
|
FBXO11
|
F-box protein 11
|
|
chr5_+_112849352
|
1.898
|
NM_022828
|
YTHDC2
|
YTH domain containing 2
|
|
chr12_+_70133179
|
1.894
|
NM_175623
NM_175625
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr18_-_45456925
|
1.871
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr11_-_3013577
|
1.865
|
NM_005969
|
NAP1L4
|
nucleosome assembly protein 1-like 4
|
|
chr21_+_38792600
|
1.865
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr15_+_91643429
|
1.864
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr14_+_103801160
|
1.860
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr12_+_70172748
|
1.844
|
NM_001024647
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr2_+_130737234
|
1.840
|
NM_032144
|
RAB6C
|
RAB6C, member RAS oncogene family
|
|
chr15_+_91073094
|
1.839
|
NM_001042574
NM_022769
|
CRTC3
|
CREB regulated transcription coactivator 3
|
|
chr17_-_78450247
|
1.831
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr3_-_184870727
|
1.808
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr7_-_87856282
|
1.798
|
NM_198901
|
SRI
|
sorcin
|
|
chr13_-_25861608
|
1.792
|
NM_004685
|
MTMR6
|
myotubularin related protein 6
|
|
chr12_-_120315074
|
1.778
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chrX_+_119738542
|
1.770
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr1_-_212965104
|
1.744
|
NM_001042549
NM_015471
|
NSL1
|
NSL1, MIND kinetochore complex component, homolog (S. cerevisiae)
|
|
chr17_+_57697036
|
1.740
|
NM_004859
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr12_-_80328926
|
1.737
|
NM_002480
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|