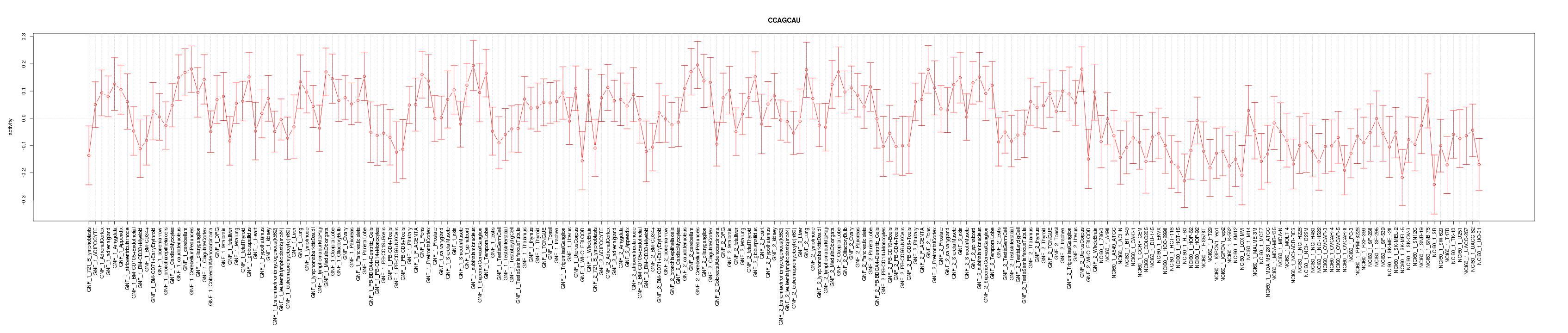
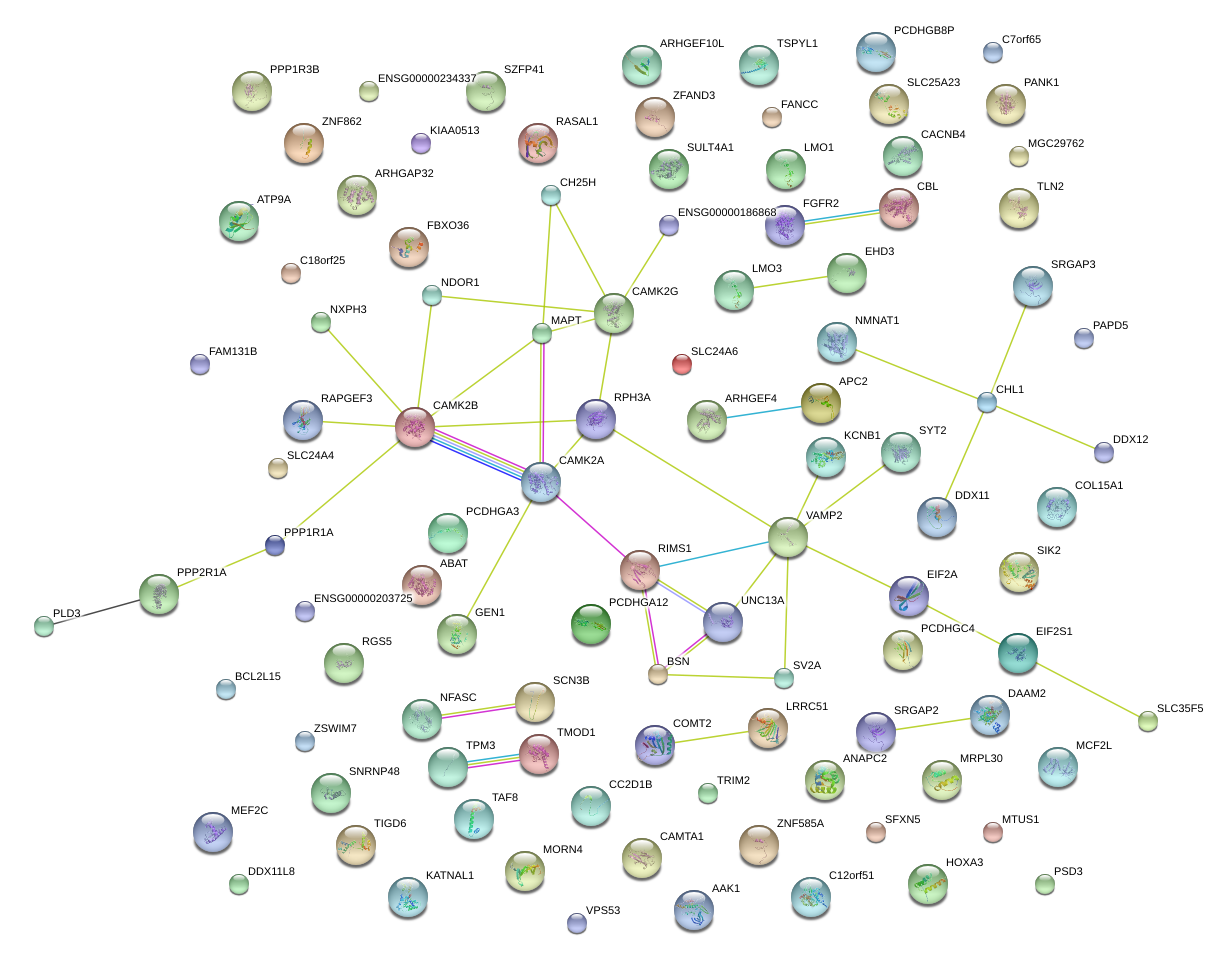
|
chr22_-_44258210
|
21.657
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr14_+_92790151
|
15.526
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr1_-_154155698
|
13.346
|
NM_001043351
NM_001043352
NM_001043353
NM_153649
|
TPM3
|
tropomyosin 3
|
|
chr14_+_92789536
|
13.105
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr1_-_149889362
|
12.534
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr5_+_140864626
|
11.435
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr5_+_140739589
|
11.047
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr17_+_43971655
|
11.020
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr10_-_91403630
|
10.911
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr3_+_49591897
|
10.525
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr17_-_15902907
|
10.151
|
NM_001042697
NM_001042698
|
ZSWIM7
|
zinc finger, SWIM-type containing 7
|
|
chr12_-_54982271
|
10.120
|
NM_006741
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A
|
|
chr8_-_18871163
|
9.824
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr14_+_92788924
|
9.823
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr5_+_140729723
|
9.807
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr5_-_88179278
|
9.615
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_+_6845381
|
9.229
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr3_+_238274
|
9.168
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr5_+_140868721
|
9.105
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr6_+_42018250
|
9.003
|
NM_138572
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr15_+_62939509
|
8.938
|
NM_015059
|
TLN2
|
talin 2
|
|
chr1_-_202679415
|
8.877
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr17_-_8066254
|
8.857
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr7_+_149535508
|
8.756
|
NM_001099220
|
ZNF862
|
zinc finger protein 862
|
|
chr11_-_123525300
|
8.673
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr5_+_140749830
|
8.671
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr16_+_50186809
|
8.629
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr1_-_202612580
|
8.458
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr5_+_140767402
|
8.121
|
NM_003736
NM_032098
|
PCDHGB4
|
protocadherin gamma subfamily B, 4
|
|
chr16_+_85061303
|
8.002
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr5_+_140797243
|
7.864
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr4_+_154074269
|
7.744
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr1_+_10003485
|
7.609
|
NM_022787
|
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1
|
|
chr8_-_18666295
|
7.514
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr5_-_149669400
|
7.430
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr20_-_50384899
|
7.320
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr11_-_128894008
|
7.279
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr3_+_150264514
|
7.186
|
NM_032025
|
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa
|
|
chr9_+_140100109
|
7.126
|
NM_001144026
NM_001144027
NM_001144028
NM_014434
|
NDOR1
|
NADPH dependent diflavin oxidoreductase 1
|
|
chr10_-_75634219
|
7.105
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr12_-_16760935
|
7.090
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_+_154125589
|
7.021
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr5_+_140810126
|
6.891
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr10_-_99393175
|
6.761
|
NM_178832
|
MORN4
|
MORN repeat containing 4
|
|
chr5_+_140753480
|
6.555
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr11_-_129062092
|
6.323
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr13_+_113623496
|
6.310
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr10_-_123353771
|
6.301
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr2_+_31456879
|
6.158
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr12_-_16759531
|
6.146
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr18_+_43753986
|
6.118
|
NM_001008239
NM_145055
|
C18orf25
|
chromosome 18 open reading frame 25
|
|
chr5_+_140777603
|
6.088
|
NM_018925
NM_032099
|
PCDHGB5
|
protocadherin gamma subfamily B, 5
|
|
chr17_+_47653176
|
6.038
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr10_-_99393851
|
6.030
|
NM_001098831
|
MORN4
|
MORN repeat containing 4
|
|
chr2_+_17935379
|
5.969
|
NM_001130009
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr2_-_152830448
|
5.910
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr9_+_101705710
|
5.828
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr12_+_113229548
|
5.748
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr2_+_17935124
|
5.714
|
NM_182625
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr11_+_71791376
|
5.696
|
NM_001145307
NM_001145308
NM_001205138
NM_145309
|
LRTOMT
|
leucine rich transmembrane and 0-methyltransferase domain containing
|
|
chr13_-_30881141
|
5.598
|
NM_001014380
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chr11_+_111473099
|
5.576
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chr9_-_98079856
|
5.554
|
NM_000136
NM_001243744
|
FANCC
|
Fanconi anemia, complementation group C
|
|
chr10_-_90967048
|
5.502
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr6_+_7590405
|
5.429
|
NM_152551
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12)
|
|
chr19_-_37701385
|
5.334
|
NM_152279
|
ZNF585B
|
zinc finger protein 585B
|
|
chr13_-_30881556
|
5.314
|
NM_032116
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chr17_-_617948
|
5.308
|
NM_001128159
NM_018289
|
VPS53
|
vacuolar protein sorting 53 homolog (S. cerevisiae)
|
|
chr1_+_17906941
|
5.216
|
NM_001011722
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr7_-_143059696
|
5.214
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr5_+_140762401
|
5.207
|
NM_018920
NM_032087
|
PCDHGA7
|
protocadherin gamma subfamily A, 7
|
|
chr2_-_69870976
|
5.161
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr8_-_17658385
|
5.115
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_+_140800536
|
5.087
|
NM_018914
NM_032091
NM_032092
|
PCDHGA11
|
protocadherin gamma subfamily A, 11
|
|
chr7_-_143059144
|
5.021
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr10_-_123356158
|
4.998
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr6_+_37787266
|
4.982
|
NM_021943
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr12_-_48152860
|
4.976
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr7_+_47694841
|
4.936
|
NM_001123065
|
C7orf65
|
chromosome 7 open reading frame 65
|
|
chr2_-_152955208
|
4.915
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr2_+_99797536
|
4.873
|
NM_145212
|
MRPL30
|
mitochondrial ribosomal protein L30
|
|
chr3_-_9291251
|
4.836
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr1_-_52831817
|
4.834
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr19_+_40854331
|
4.741
|
NM_001031696
NM_012268
|
PLD3
|
phospholipase D family, member 3
|
|
chr1_+_17866276
|
4.655
|
NM_018125
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr2_-_73298815
|
4.630
|
NM_144579
|
SFXN5
|
sideroflexin 5
|
|
chr5_+_140771482
|
4.588
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chr16_+_8768403
|
4.483
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr12_-_113772900
|
4.454
|
NM_024959
|
SLC24A6
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 6
|
|
chr1_+_204913353
|
4.451
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr2_+_131674223
|
4.424
|
NM_015320
NM_032995
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4
|
|
chr5_+_140718353
|
4.393
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr2_-_152955502
|
4.331
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr5_+_140734767
|
4.316
|
NM_018917
NM_032053
|
PCDHGA4
|
protocadherin gamma subfamily A, 4
|
|
chr1_+_204797774
|
4.315
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr8_-_17579729
|
4.276
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr9_+_100263461
|
4.272
|
NM_001166116
|
TMOD1
|
tropomodulin 1
|
|
chr5_-_149380729
|
4.251
|
NM_030953
|
TIGD6
|
tigger transposable element derived 6
|
|
chr19_-_37263653
|
4.217
|
NM_001193552
|
ZNF850
|
zinc finger protein 850
|
|
chr6_-_116601102
|
4.185
|
NM_003309
|
TSPYL1
|
TSPY-like 1
|
|
chr19_-_17799005
|
4.169
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr11_+_119076974
|
4.147
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr1_-_163172624
|
4.137
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr19_+_1450117
|
4.122
|
NM_005883
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr19_+_52693054
|
4.113
|
NM_014225
|
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha
|
|
chr7_-_27159213
|
4.112
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr10_-_123357503
|
4.108
|
NM_000141
NM_001144917
NM_001144918
NM_001144919
NM_022970
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr2_-_114514196
|
4.103
|
NM_025181
|
SLC35F5
|
solute carrier family 35, member F5
|
|
chr12_-_113573267
|
4.101
|
NM_001193521
|
RASAL1
|
RAS protein activator like 1 (GAP1 like)
|
|
chr8_-_9009050
|
4.101
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr5_+_140743755
|
4.101
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr12_-_112819755
|
4.092
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr1_-_114430168
|
4.049
|
NM_001010922
|
BCL2L15
|
BCL2-like 15
|
|
chr2_+_230787195
|
4.040
|
NM_174899
|
FBXO36
|
F-box protein 36
|
|
chr9_+_100263818
|
4.032
|
NM_003275
|
TMOD1
|
tropomodulin 1
|
|
chr1_-_163172863
|
4.028
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr20_-_48099178
|
4.024
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr6_+_72922513
|
4.016
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr12_-_113574020
|
3.979
|
NM_001193520
NM_004658
|
RASAL1
|
RAS protein activator like 1 (GAP1 like)
|
|
chr6_+_39760148
|
3.963
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr10_-_118501981
|
3.887
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr5_+_140855543
|
3.864
|
NM_002588
NM_032402
NM_032403
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr22_+_32340435
|
3.849
|
NM_003405
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr16_-_51184507
|
3.835
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr19_-_6459735
|
3.793
|
NM_024103
|
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23
|
|
chr6_+_36853639
|
3.790
|
NM_152734
|
C6orf89
|
chromosome 6 open reading frame 89
|
|
chr5_-_149535420
|
3.789
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr7_+_30960914
|
3.774
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr1_+_54519259
|
3.763
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr15_+_80696569
|
3.756
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr12_-_48152180
|
3.743
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_57113292
|
3.729
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr20_-_41818464
|
3.727
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr7_-_51384458
|
3.715
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr13_+_113656027
|
3.684
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr1_+_169075869
|
3.678
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr8_+_21915371
|
3.630
|
NM_001114135
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr2_+_26403584
|
3.629
|
NM_001191033
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr22_-_20104745
|
3.613
|
NM_022727
NM_182984
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr15_-_33360084
|
3.603
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr7_+_45614081
|
3.542
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr12_-_57400199
|
3.540
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr3_-_57199402
|
3.540
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr3_-_13114547
|
3.511
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr11_+_133938819
|
3.507
|
NM_001205329
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr5_-_443168
|
3.488
|
NM_138464
|
C5orf55
|
chromosome 5 open reading frame 55
|
|
chr9_-_95432542
|
3.485
|
NM_022755
|
IPPK
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase
|
|
chr20_+_37434294
|
3.399
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr17_+_40118696
|
3.381
|
NM_033133
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chr9_+_17578952
|
3.381
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr5_-_64777703
|
3.375
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr19_+_32896515
|
3.347
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr2_+_241375030
|
3.309
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr9_-_34710062
|
3.290
|
NM_002989
|
CCL21
|
chemokine (C-C motif) ligand 21
|
|
chr7_+_156742402
|
3.288
|
NM_138400
|
NOM1
|
nucleolar protein with MIF4G domain 1
|
|
chr1_-_203320287
|
3.283
|
NM_002023
|
FMOD
|
fibromodulin
|
|
chr16_+_5008317
|
3.283
|
NM_014692
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae)
|
|
chr7_-_27166638
|
3.252
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr3_-_44519156
|
3.243
|
NM_181489
|
ZNF445
|
zinc finger protein 445
|
|
chr2_+_46524448
|
3.232
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr5_+_140723600
|
3.207
|
NM_018916
NM_032011
|
PCDHGA3
|
protocadherin gamma subfamily A, 3
|
|
chr16_+_89894853
|
3.201
|
NM_032451
|
SPIRE2
|
spire homolog 2 (Drosophila)
|
|
chr15_-_45815001
|
3.191
|
NM_013309
|
SLC30A4
|
solute carrier family 30 (zinc transporter), member 4
|
|
chr19_+_54466289
|
3.188
|
NM_031895
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8
|
|
chr9_+_133884503
|
3.153
|
NM_006059
|
LAMC3
|
laminin, gamma 3
|
|
chr6_+_72926144
|
3.128
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr7_+_30951414
|
3.120
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr1_-_36235422
|
3.109
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr1_+_164528586
|
3.105
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr18_-_53255734
|
3.102
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr16_+_8806825
|
3.093
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr8_+_21911062
|
3.091
|
NM_001114137
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr9_+_140033588
|
3.066
|
NM_000832
NM_001185090
NM_001185091
NM_007327
NM_021569
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr5_+_140792507
|
3.061
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chrX_+_70586113
|
3.041
|
NM_004606
NM_138923
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa
|
|
chr19_+_54024176
|
3.029
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr1_-_160990971
|
2.997
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr9_+_139560193
|
2.990
|
NM_201446
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr3_+_13521714
|
2.986
|
NM_024827
NM_001136041
|
HDAC11
|
histone deacetylase 11
|
|
chr2_+_7057471
|
2.948
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr12_+_4918341
|
2.941
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr6_+_73076431
|
2.935
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr21_+_35747748
|
2.924
|
NM_058182
|
FAM165B
|
family with sequence similarity 165, member B
|
|
chr7_-_158622202
|
2.897
|
NM_020728
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr15_+_43809805
|
2.868
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr7_-_108096693
|
2.867
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr17_+_18163836
|
2.860
|
NM_001144900
NM_139162
|
SMCR7
|
Smith-Magenis syndrome chromosome region, candidate 7
|
|
chr1_+_226736500
|
2.857
|
NM_001003665
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr9_-_91793681
|
2.855
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr12_-_48551269
|
2.837
|
NM_024095
|
ASB8
|
ankyrin repeat and SOCS box containing 8
|
|
chr22_-_44708730
|
2.832
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr19_-_6110561
|
2.821
|
NM_000635
NM_134433
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression)
|
|
chr8_+_21912327
|
2.800
|
NM_001114136
NM_001114139
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr17_-_18585571
|
2.792
|
NM_001145045
|
ZNF286B
|
zinc finger protein 286B
|
|
chr8_-_33424642
|
2.784
|
NM_024787
|
RNF122
|
ring finger protein 122
|
|
chr17_-_62207475
|
2.773
|
NM_001433
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1
|
|
chr3_-_13008959
|
2.773
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr1_+_184356128
|
2.762
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr1_+_16062808
|
2.755
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr7_-_5553305
|
2.753
|
NM_024963
|
FBXL18
|
F-box and leucine-rich repeat protein 18
|