|
chr6_+_43543877
|
80.696
|
NM_006502
|
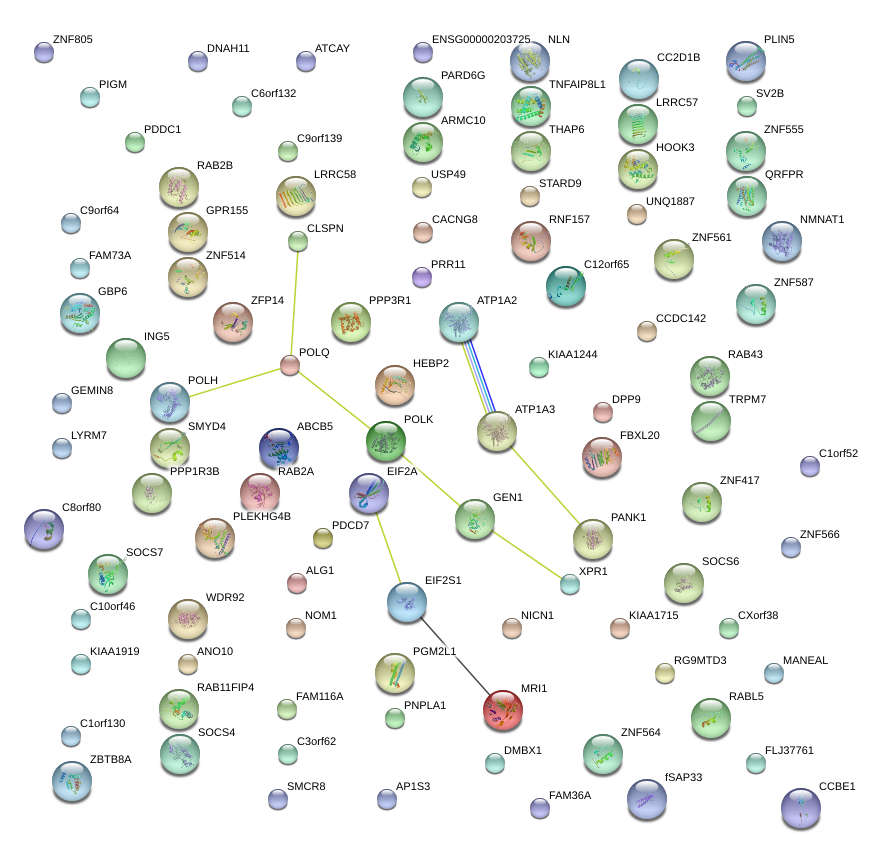
POLH
|
polymerase (DNA directed), eta
|
|
chr17_-_74236272
|
76.308
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr11_-_777470
|
74.066
|
NM_182612
|
PDDC1
|
Parkinson disease 7 domain containing 1
|
|
chr1_+_24882565
|
72.735
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr2_-_74710356
|
65.897
|
NM_032779
|
CCDC142
|
coiled-coil domain containing 142
|
|
chr2_+_17935379
|
64.819
|
NM_001130009
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr2_-_68384575
|
64.730
|
NM_138458
|
WDR92
|
WD repeat domain 92
|
|
chr2_+_242641310
|
64.317
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr17_-_1733083
|
64.158
|
NM_052928
|
SMYD4
|
SET and MYND domain containing 4
|
|
chr19_-_36870064
|
63.633
|
NM_020917
|
ZFP14
|
zinc finger protein 14 homolog (mouse)
|
|
chr2_+_17935124
|
63.186
|
NM_182625
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr1_-_36235422
|
62.197
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr15_+_91643429
|
60.793
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr2_-_175351755
|
58.569
|
NM_001033045
NM_152529
|
GPR155
|
G protein-coupled receptor 155
|
|
chr8_-_27941387
|
57.271
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr19_+_52956758
|
57.056
|
NM_001099694
|
ZNF578
|
zinc finger protein 578
|
|
chr19_+_4639526
|
55.453
|
NM_152362
|
TNFAIP8L1
|
tumor necrosis factor, alpha-induced protein 8-like 1
|
|
chr8_+_42752019
|
54.886
|
NM_032410
|
HOOK3
|
hook homolog 3 (Drosophila)
|
|
chr19_+_54466289
|
53.317
|
NM_031895
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8
|
|
chr1_+_10003485
|
53.133
|
NM_022787
|
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1
|
|
chr15_+_42867708
|
52.785
|
NM_020759
|
STARD9
|
StAR-related lipid transfer (START) domain containing 9
|
|
chr5_+_130506635
|
52.552
|
NM_181705
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chrX_-_40506800
|
52.549
|
NM_144970
|
CXorf38
|
chromosome X open reading frame 38
|
|
chr3_-_57678594
|
52.545
|
NM_152678
|
FAM116A
|
family with sequence similarity 116, member A
|
|
chr15_+_91643181
|
52.362
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr1_+_46972667
|
52.014
|
NM_147192
NM_172225
|
DMBX1
|
diencephalon/mesencephalon homeobox 1
|
|
chr1_+_33004771
|
51.767
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr3_-_120068077
|
51.300
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr1_+_78245306
|
51.075
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr6_+_138483024
|
49.366
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr4_+_76439653
|
49.299
|
NM_144721
|
THAP6
|
THAP domain containing 6
|
|
chr3_-_128879873
|
49.002
|
NM_001204890
NM_001199469
NM_020701
|
ISY1-RAB43
ISY1
|
ISY1-RAB43 readthrough
ISY1 splicing factor homolog (S. cerevisiae)
|
|
chr19_-_9731883
|
48.879
|
NM_152289
|
ZNF561
|
zinc finger protein 561
|
|
chr8_-_9009050
|
48.650
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr17_+_18218569
|
47.723
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr7_+_102715293
|
47.309
|
NM_001161009
NM_001161010
NM_001161011
NM_001161012
NM_001161013
NM_031905
|
ARMC10
|
armadillo repeat containing 10
|
|
chr12_-_121342050
|
46.720
|
NM_139015
|
SPPL3
|
signal peptide peptidase-like 3
|
|
chr3_+_150264514
|
46.285
|
NM_032025
|
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa
|
|
chr4_-_122302112
|
45.761
|
NM_198179
|
QRFPR
|
pyroglutamylated RFamide peptide receptor
|
|
chr5_+_65017993
|
45.744
|
NM_020726
|
NLN
|
neurolysin (metallopeptidase M3 family)
|
|
chr1_+_160085519
|
45.727
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr1_+_228353428
|
45.486
|
NM_001010867
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae)
|
|
chr6_-_41863086
|
45.352
|
NM_018561
|
USP49
|
ubiquitin specific peptidase 49
|
|
chr17_+_29718641
|
45.130
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr1_+_54519259
|
44.958
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr1_-_52831817
|
44.880
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr1_-_160001723
|
43.508
|
NM_145167
|
PIGM
|
phosphatidylinositol glycan anchor biosynthesis, class M
|
|
chr15_-_50978983
|
43.282
|
NM_017672
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr1_+_180601121
|
42.932
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr10_-_91403630
|
42.858
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr9_+_37753769
|
42.680
|
NM_144964
|
RG9MTD3
|
RNA (guanine-9-) methyltransferase domain containing 3
|
|
chr2_-_176866925
|
42.352
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chr5_+_140372
|
42.180
|
NM_052909
|
PLEKHG4B
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B
|
|
chr7_+_20655244
|
42.141
|
NM_001163941
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr19_+_13875299
|
41.754
|
NM_001031727
NM_032285
|
MRI1
|
methylthioribose-1-phosphate isomerase homolog (S. cerevisiae)
|
|
chr19_-_36980336
|
41.370
|
NM_032838
NM_001145343
NM_001145344
NM_001145345
|
ZNF566
|
zinc finger protein 566
|
|
chr17_+_36507708
|
41.204
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr14_-_21945031
|
41.176
|
NM_032846
|
RAB2B
|
RAB2B, member RAS oncogene family
|
|
chr3_-_49314281
|
40.783
|
NM_198562
|
C3orf62
|
chromosome 3 open reading frame 62
|
|
chr17_+_57232859
|
40.453
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr6_+_111580481
|
40.160
|
NM_153369
|
KIAA1919
|
KIAA1919
|
|
chr20_+_54933982
|
40.108
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr14_-_21944826
|
39.898
|
NM_001163380
|
RAB2B
|
RAB2B, member RAS oncogene family
|
|
chr3_-_49466744
|
39.593
|
NM_032316
|
NICN1
|
nicolin 1
|
|
chr22_-_39052579
|
39.419
|
NM_001013647
|
LOC646851
|
putative uncharacterized protein LOC388900
|
|
chr17_-_37557851
|
39.192
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr9_-_86571652
|
39.151
|
NM_032307
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr15_-_42840992
|
38.757
|
NM_153260
|
LRRC57
|
leucine rich repeat containing 57
|
|
chr1_-_85725315
|
37.804
|
NM_198077
|
C1orf52
|
chromosome 1 open reading frame 52
|
|
chr6_-_42110714
|
37.783
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr7_+_156742402
|
37.604
|
NM_138400
|
NOM1
|
nucleolar protein with MIF4G domain 1
|
|
chr19_+_2841432
|
37.193
|
NM_001172775
NM_152791
|
ZNF555
|
zinc finger protein 555
|
|
chr1_+_89829435
|
37.158
|
NM_198460
|
GBP6
|
guanylate binding protein family, member 6
|
|
chr2_-_95825262
|
36.920
|
NM_032788
|
ZNF514
|
zinc finger protein 514
|
|
chr1_+_38261079
|
36.493
|
NM_152496
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr2_-_224702300
|
36.273
|
NM_001039569
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr19_+_3880617
|
35.882
|
NM_033064
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chr9_+_139921915
|
35.532
|
NM_207511
|
C9orf139
|
chromosome 9 open reading frame 139
|
|
chr6_+_36210944
|
35.484
|
NM_173676
NM_001145716
|
PNPLA1
|
patatin-like phospholipase domain containing 1
|
|
chr19_-_4535202
|
35.088
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr12_+_123717843
|
35.052
|
NM_152269
NM_001194995
|
C12orf65
|
chromosome 12 open reading frame 65
|
|
chr2_-_200820458
|
34.828
|
NM_001039693
|
TYW5
|
tRNA-yW synthesizing protein 5
|
|
chr11_-_65547821
|
34.703
|
NM_138368
|
DKFZp761E198
|
adaptor protein 5
|
|
chr11_+_17373138
|
34.639
|
NM_001202439
|
B7H6
|
B7 homolog 6
|
|
chr10_-_120514669
|
34.469
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr11_-_74109417
|
34.416
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chrX_-_14047958
|
34.366
|
NM_001042479
NM_001042480
NM_017856
|
GEMIN8
|
gem (nuclear organelle) associated protein 8
|
|
chr19_+_57752051
|
34.222
|
NM_001023563
NM_001145078
|
ZNF805
|
zinc finger protein 805
|
|
chr18_-_57364643
|
34.041
|
NM_133459
|
CCBE1
|
collagen and calcium binding EGF domains 1
|
|
chr19_-_58427934
|
33.839
|
NM_152475
|
ZNF417
|
zinc finger protein 417
|
|
chr16_+_5121809
|
33.777
|
NM_019109
|
ALG1
|
asparagine-linked glycosylation 1, beta-1,4-mannosyltransferase homolog (S. cerevisiae)
|
|
chr19_-_4723814
|
33.683
|
NM_139159
|
DPP9
|
dipeptidyl-peptidase 9
|
|
chr7_-_100964923
|
33.384
|
NM_001130820
NM_001130821
NM_001130822
NM_022777
|
RABL5
|
RAB, member RAS oncogene family-like 5
|
|
chr18_-_78005230
|
33.281
|
NM_032510
|
PARD6G
|
par-6 partitioning defective 6 homolog gamma (C. elegans)
|
|
chr19_-_12662191
|
33.217
|
NM_144976
|
ZNF564
|
zinc finger protein 564
|
|
chr15_-_65425650
|
33.150
|
NM_005707
|
PDCD7
|
programmed cell death 7
|
|
chr1_+_244998592
|
33.142
|
NM_198076
|
FAM36A
|
family with sequence similarity 36, member A
|
|
chr15_+_59063389
|
32.987
|
NM_001040450
NM_001040453
|
FAM63B
|
family with sequence similarity 63, member B
|
|
chr1_-_151882360
|
32.909
|
NM_053055
|
THEM4
|
thioesterase superfamily member 4
|
|
chr19_+_32836500
|
32.866
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr5_-_118324223
|
32.776
|
NM_173666
|
DTWD2
|
DTW domain containing 2
|
|
chr1_+_38259458
|
32.485
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr7_+_4721722
|
32.472
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr1_+_15573767
|
32.468
|
NM_052929
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1
|
|
chr10_+_38299525
|
32.447
|
NM_006954
NM_006974
|
ZNF33A
|
zinc finger protein 33A
|
|
chr4_+_40058507
|
32.433
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr10_+_106034886
|
32.378
|
NM_001191014
NM_001191015
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr3_+_40566341
|
32.282
|
NM_001098414
NM_198484
|
ZNF621
|
zinc finger protein 621
|
|
chr8_-_127570710
|
32.210
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr17_+_43971655
|
32.086
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr19_-_18335302
|
31.712
|
NM_001098819
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific
|
|
chr14_+_74111577
|
31.552
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr6_+_36238236
|
31.551
|
NM_001145717
|
PNPLA1
|
patatin-like phospholipase domain containing 1
|
|
chr8_-_9008138
|
31.505
|
NM_024607
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr14_+_74551655
|
31.415
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr3_-_141944441
|
31.376
|
NM_001039547
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr1_-_35325335
|
31.364
|
NM_001164825
NM_138428
|
C1orf212
|
chromosome 1 open reading frame 212
|
|
chr12_+_32654976
|
31.307
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr3_-_15106806
|
30.894
|
NM_022497
|
MRPS25
|
mitochondrial ribosomal protein S25
|
|
chr2_-_62081049
|
30.857
|
NM_001201543
NM_032180
|
FAM161A
|
family with sequence similarity 161, member A
|
|
chr4_+_48833041
|
30.468
|
NM_001079840
NM_001079841
NM_017830
NM_001079839
NM_001079842
NM_001168254
|
OCIAD1
|
OCIA domain containing 1
|
|
chr2_-_220110116
|
30.456
|
NM_024506
|
GLB1L
|
galactosidase, beta 1-like
|
|
chr1_-_54665670
|
30.223
|
NM_001031672
|
CYB5RL
|
cytochrome b5 reductase-like
|
|
chr7_-_102105307
|
30.049
|
NM_017621
|
ALKBH4
|
alkB, alkylation repair homolog 4 (E. coli)
|
|
chr17_-_26941178
|
29.702
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr16_+_50186809
|
29.462
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr3_-_194991778
|
29.450
|
NM_152531
|
XXYLT1
|
xyloside xylosyltransferase 1
|
|
chr1_+_19638739
|
29.405
|
NM_001040125
NM_001040126
NM_017765
|
PQLC2
|
PQ loop repeat containing 2
|
|
chr4_-_73935408
|
29.348
|
NM_173827
|
COX18
|
COX18 cytochrome c oxidase assembly homolog (S. cerevisiae)
|
|
chr19_-_51920841
|
29.334
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr19_+_39881950
|
29.304
|
NM_017592
|
MED29
|
mediator complex subunit 29
|
|
chr3_-_15140652
|
29.278
|
NM_022340
|
ZFYVE20
|
zinc finger, FYVE domain containing 20
|
|
chr11_+_76745434
|
29.157
|
NM_138706
|
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase)
|
|
chr19_-_18337234
|
29.086
|
NM_001098818
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific
|
|
chr11_-_102962903
|
29.045
|
NM_032299
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae)
|
|
chr19_+_52772823
|
28.802
|
NM_001010851
|
ZNF766
|
zinc finger protein 766
|
|
chr6_-_121655606
|
28.749
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr7_-_766875
|
28.682
|
NM_001164760
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr19_+_37997840
|
28.533
|
NM_001013659
|
ZNF793
|
zinc finger protein 793
|
|
chr3_-_126076057
|
28.469
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr2_+_99797536
|
28.440
|
NM_145212
|
MRPL30
|
mitochondrial ribosomal protein L30
|
|
chr16_-_23724820
|
28.407
|
NM_033266
|
ERN2
|
endoplasmic reticulum to nucleus signaling 2
|
|
chr7_+_138482696
|
28.294
|
NM_001085429
|
TMEM213
|
transmembrane protein 213
|
|
chr20_-_47894591
|
28.268
|
NM_021035
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr17_+_74733468
|
28.169
|
NM_001242532
NM_001242533
NM_001242535
NM_001242536
NM_001242537
NM_024311
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr19_+_18263998
|
28.083
|
NM_005027
|
PIK3R2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta)
|
|
chr1_+_53308182
|
27.837
|
NM_001004339
|
ZYG11A
|
zyg-11 homolog A (C. elegans)
|
|
chr8_-_96281399
|
27.801
|
NM_177965
|
C8orf37
|
chromosome 8 open reading frame 37
|
|
chr18_-_21017816
|
27.768
|
NM_032933
|
TMEM241
|
transmembrane protein 241
|
|
chr9_-_115480363
|
27.495
|
NM_021218
|
C9orf80
|
chromosome 9 open reading frame 80
|
|
chr19_-_16683188
|
27.441
|
NM_024881
|
SLC35E1
|
solute carrier family 35, member E1
|
|
chrX_+_13671224
|
27.339
|
NM_152634
|
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing
|
|
chr15_-_81282134
|
27.164
|
NM_015154
|
MESDC2
|
mesoderm development candidate 2
|
|
chr15_+_42840991
|
27.145
|
NM_001130447
NM_018097
|
HAUS2
|
HAUS augmin-like complex, subunit 2
|
|
chr5_-_10308167
|
27.043
|
NM_138809
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas)
|
|
chr3_+_93698959
|
26.970
|
NM_001174150
NM_001174151
NM_144996
NM_182896
|
ARL13B
|
ADP-ribosylation factor-like 13B
|
|
chr2_-_208890207
|
26.700
|
NM_001080475
|
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3
|
|
chr12_+_175872
|
26.686
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr16_+_18995255
|
26.635
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr10_+_134145639
|
26.635
|
NM_001143757
|
LRRC27
|
leucine rich repeat containing 27
|
|
chr1_-_35324645
|
26.558
|
NM_001164824
|
C1orf212
|
chromosome 1 open reading frame 212
|
|
chr9_-_4741267
|
26.552
|
NM_001199852
NM_016282
|
AK3
|
adenylate kinase 3
|
|
chr12_-_66524501
|
26.550
|
NM_032338
|
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia)
|
|
chrX_-_1655999
|
26.531
|
NM_178129
|
P2RY8
CRLF2
|
purinergic receptor P2Y, G-protein coupled, 8
cytokine receptor-like factor 2
|
|
chrY_-_1605999
|
26.531
|
NM_178129
|
P2RY8
CRLF2
|
purinergic receptor P2Y, G-protein coupled, 8
cytokine receptor-like factor 2
|
|
chr20_-_62205559
|
26.511
|
NM_001037335
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chr1_+_169337193
|
26.511
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr3_+_42947649
|
26.484
|
NM_001134656
|
ZNF662
|
zinc finger protein 662
|
|
chr9_-_4741798
|
26.249
|
NM_001199855
|
AK3
|
adenylate kinase 3
|
|
chrX_-_50557019
|
26.242
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr16_+_22825806
|
26.103
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr5_+_94890777
|
26.077
|
NM_198150
|
ARSK
|
arylsulfatase family, member K
|
|
chr18_-_14132426
|
25.942
|
NM_145287
|
ZNF519
|
zinc finger protein 519
|
|
chr19_-_3869012
|
25.921
|
NM_001145640
NM_015174
|
ZFR2
|
zinc finger RNA binding protein 2
|
|
chr7_-_767286
|
25.898
|
NM_001164758
NM_001164759
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr1_-_173793776
|
25.882
|
NM_001127181
NM_033319
|
CENPL
|
centromere protein L
|
|
chrX_-_15333726
|
25.833
|
NM_001201583
NM_080873
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr5_+_54455945
|
25.813
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr1_-_173793268
|
25.706
|
NM_001171182
|
CENPL
|
centromere protein L
|
|
chr1_-_100715388
|
25.696
|
NM_001918
|
DBT
|
dihydrolipoamide branched chain transacylase E2
|
|
chr19_-_57678855
|
25.684
|
NM_001012729
|
DUXA
|
double homeobox A
|
|
chr9_-_100954883
|
25.657
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr2_-_68694389
|
25.628
|
NM_001024680
|
FBXO48
|
F-box protein 48
|
|
chr1_-_62785067
|
25.528
|
NM_181712
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chr1_-_202679415
|
25.495
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr19_+_58095507
|
25.453
|
NM_001010879
|
ZIK1
|
zinc finger protein interacting with K protein 1 homolog (mouse)
|
|
chr17_-_5372235
|
25.301
|
NM_001199699
NM_020162
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33
|
|
chr17_-_8301143
|
25.295
|
NM_001146684
|
RNF222
|
ring finger protein 222
|
|
chr14_+_23299083
|
25.091
|
NM_178336
NM_180982
NM_181304
NM_181305
NM_181306
NM_181307
|
MRPL52
|
mitochondrial ribosomal protein L52
|
|
chr6_+_36410543
|
25.087
|
NM_173562
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr3_-_120461362
|
25.010
|
NM_173825
|
RABL3
|
RAB, member of RAS oncogene family-like 3
|
|
chr17_-_18950238
|
25.008
|
NM_006613
|
GRAP
|
GRB2-related adaptor protein
|
|
chr12_+_123718290
|
24.986
|
NM_001143905
|
C12orf65
|
chromosome 12 open reading frame 65
|
|
chr6_+_99968851
|
24.983
|
NM_001195131
|
LOC100130890
|
uncharacterized LOC100130890
|
|
chr19_-_37701385
|
24.954
|
NM_152279
|
ZNF585B
|
zinc finger protein 585B
|
|
chr10_-_79789290
|
24.828
|
NM_007055
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa
|
|
chr10_-_75910525
|
24.808
|
NM_012095
NM_207012
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit
|
|
chr10_+_18429605
|
24.631
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr19_-_5340700
|
24.620
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr9_-_4741085
|
24.487
|
NM_001199854
|
AK3
|
adenylate kinase 3
|