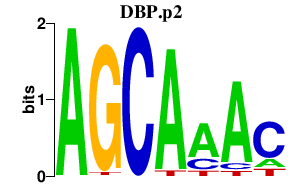
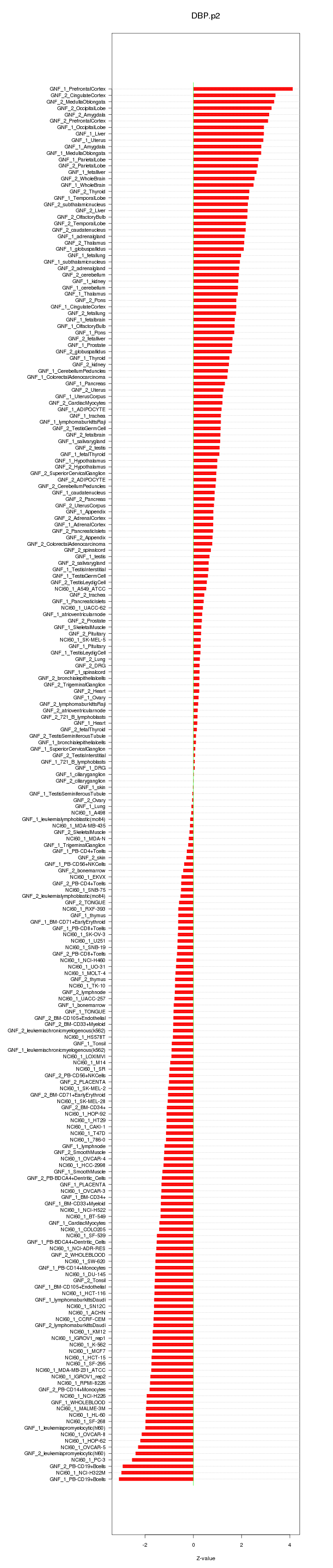
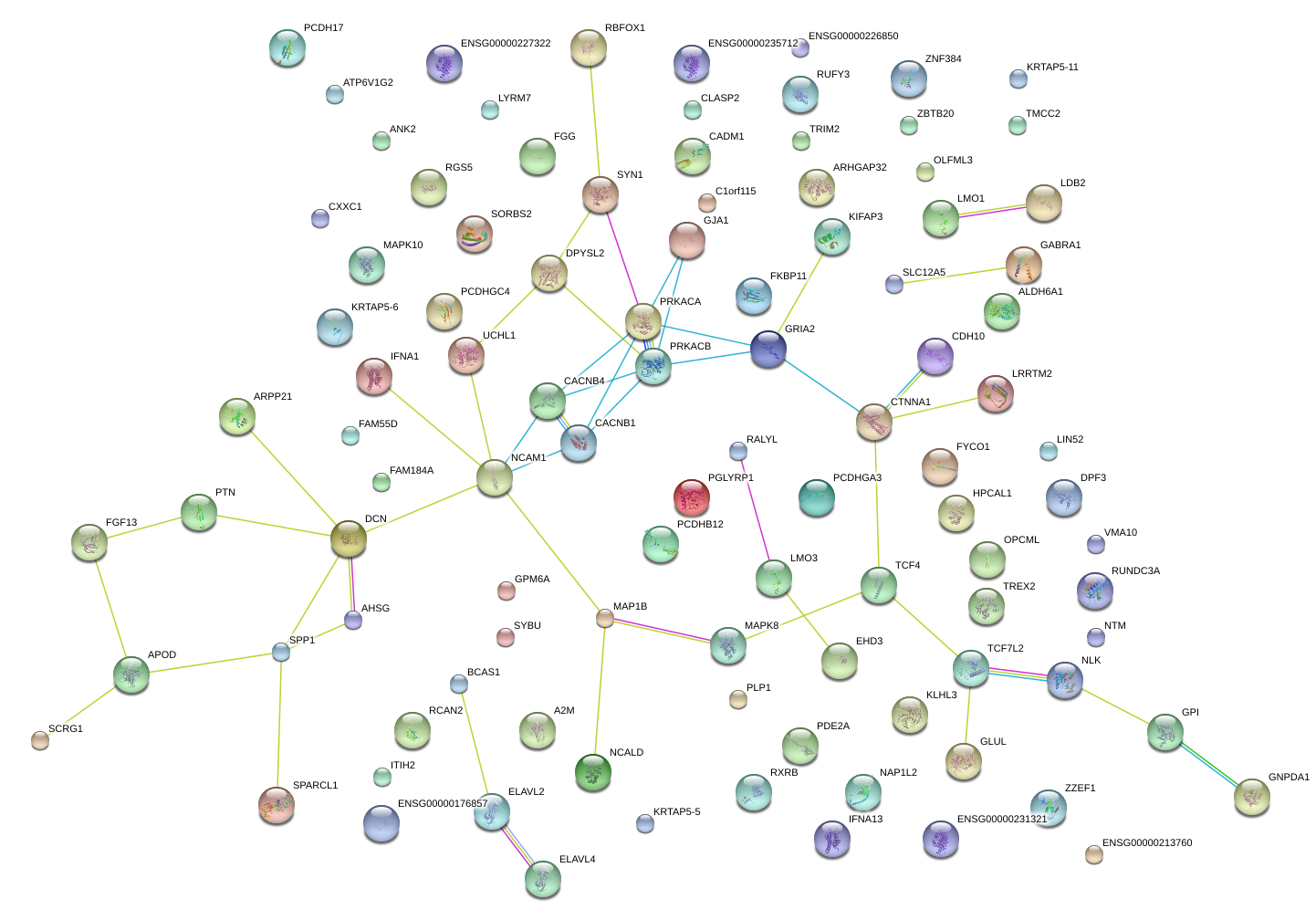
|
chr4_-_176733413
|
43.745
|
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_87281177
|
30.826
|
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr5_+_71502700
|
30.744
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr4_-_186732208
|
30.631
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_-_16759733
|
25.826
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_138210992
|
25.175
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr12_-_16759939
|
24.182
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_-_91576579
|
23.628
|
|
DCN
|
decorin
|
|
chr4_-_186732047
|
23.081
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_+_154125589
|
22.517
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr8_-_110693233
|
21.722
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr4_-_176733901
|
21.083
|
|
GPM6A
|
glycoprotein M6A
|
|
chrX_-_137821514
|
20.858
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr3_-_114343791
|
20.778
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr4_-_87281374
|
20.300
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr5_+_161277391
|
20.244
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr4_-_88450601
|
19.991
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr8_-_110655418
|
19.856
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr6_-_31514393
|
19.473
|
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr4_+_158141735
|
19.469
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr5_-_138211054
|
19.386
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr12_-_6798419
|
19.349
|
|
ZNF384
|
zinc finger protein 384
|
|
chr12_-_16759531
|
19.197
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_-_152830448
|
18.943
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr1_+_84609941
|
17.953
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr4_-_174320616
|
17.825
|
NM_007281
|
SCRG1
|
stimulator of chondrogenesis 1
|
|
chr6_-_31514620
|
17.521
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr20_+_44650260
|
17.312
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr6_-_46293628
|
17.210
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr8_-_102803438
|
16.850
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr1_-_163113938
|
16.538
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr4_-_88450243
|
16.378
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr4_-_16900256
|
15.677
|
|
LDB2
|
LIM domain binding 2
|
|
chr12_-_16759430
|
15.627
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_-_16900348
|
15.536
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_16900028
|
15.490
|
|
LDB2
|
LIM domain binding 2
|
|
chr8_-_102803196
|
15.214
|
|
NCALD
|
neurocalcin delta
|
|
chr5_-_42811959
|
15.072
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr8_-_102803178
|
15.041
|
|
NCALD
|
neurocalcin delta
|
|
chr6_-_46293404
|
14.889
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr5_-_24645084
|
14.139
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr12_-_91576805
|
13.983
|
NM_001920
|
DCN
|
decorin
|
|
chr10_+_7745341
|
13.751
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr4_+_71587674
|
13.749
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr14_-_74551073
|
13.447
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr11_-_114466471
|
13.018
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr6_-_33168448
|
12.969
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr11_-_72380107
|
12.766
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr13_+_58205788
|
12.754
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr11_-_128894008
|
12.536
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr1_-_182354649
|
12.496
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_+_140588267
|
12.285
|
NM_018932
|
PCDHB12
|
protocadherin beta 12
|
|
chr12_-_9268513
|
12.154
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr11_+_112831968
|
12.105
|
NM_000615
NM_001076682
NM_001242607
NM_001242608
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr10_+_7745284
|
12.047
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chrX_-_47479229
|
11.881
|
NM_006950
NM_133499
|
SYN1
|
synapsin I
|
|
chr4_+_113970784
|
11.774
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chrX_-_72434656
|
11.756
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr5_+_140739589
|
11.733
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr12_-_6798528
|
11.733
|
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr14_-_74551159
|
11.655
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr12_-_9268522
|
11.507
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr20_-_52687303
|
11.470
|
NM_003657
|
BCAS1
|
breast carcinoma amplified sequence 1
|
|
chr1_+_114522029
|
11.314
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr8_+_85095501
|
11.237
|
NM_173848
|
RALYL
|
RALY RNA binding protein-like
|
|
chr4_+_41258941
|
11.217
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr11_-_133402227
|
11.215
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr12_-_9268440
|
11.019
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr14_-_73360792
|
10.954
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chrX_-_72434623
|
10.941
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr5_+_140797243
|
10.926
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr5_+_130506656
|
10.859
|
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chr3_+_35721115
|
10.597
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr16_+_7382750
|
10.487
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr6_+_121756744
|
10.451
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr3_-_33700480
|
10.443
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr6_+_30594607
|
10.294
|
NM_024909
NM_001031722
NM_001190724
|
ATAT1
|
alpha tubulin acetyltransferase 1
|
|
chrX_+_103031433
|
10.214
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr13_+_58205943
|
10.128
|
|
PCDH17
|
protocadherin 17
|
|
chr6_-_119470336
|
10.124
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr5_+_140864626
|
10.102
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr4_+_88896868
|
9.840
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_+_31456879
|
9.829
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr7_-_137028433
|
9.803
|
|
PTN
|
pleiotrophin
|
|
chr1_+_205236591
|
9.753
|
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr4_+_88896838
|
9.734
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_-_155533882
|
9.707
|
NM_000509
NM_021870
|
FGG
|
fibrinogen gamma chain
|
|
chr8_+_26435920
|
9.696
|
NM_001244604
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr11_+_112832201
|
9.445
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr17_+_42385780
|
9.435
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr9_-_21368005
|
9.366
|
NM_006900
|
IFNA1
IFNA13
|
interferon, alpha 1
interferon, alpha 13
|
|
chr5_-_137071462
|
9.328
|
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr11_-_114466397
|
9.306
|
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr14_+_74551655
|
9.138
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr11_-_66639602
|
9.102
|
|
|
|
|
chr1_+_50574589
|
9.092
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr17_+_26369617
|
9.074
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr11_-_115088628
|
9.070
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr1_+_220863627
|
8.991
|
NM_024709
|
C1orf115
|
chromosome 1 open reading frame 115
|
|
chr11_+_1718424
|
8.912
|
NM_001012416
|
KRTAP5-6
|
keratin associated protein 5-6
|
|
chr1_+_84630374
|
8.900
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr7_-_28998028
|
8.865
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr1_-_170038055
|
8.855
|
NM_001204517
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr12_-_9268475
|
8.781
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr12_-_6798619
|
8.746
|
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr18_-_53303129
|
8.740
|
NM_001243226
|
TCF4
|
transcription factor 4
|
|
chrX_-_72434667
|
8.701
|
NM_021963
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr3_-_195310770
|
8.659
|
|
APOD
|
apolipoprotein D
|
|
chr8_+_85095702
|
8.626
|
|
RALYL
|
RALY RNA binding protein-like
|
|
chr3_+_186330711
|
8.540
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr3_+_35722428
|
8.473
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr7_-_137028275
|
8.436
|
|
PTN
|
pleiotrophin
|
|
chr6_-_46459710
|
8.391
|
NM_001251973
|
RCAN2
|
regulator of calcineurin 2
|
|
chr9_-_91793681
|
8.352
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr3_-_114343052
|
8.305
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr4_+_47033294
|
8.265
|
NM_000812
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1
|
|
chr1_-_241520384
|
8.203
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr1_+_183774214
|
8.166
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr5_+_31799030
|
8.163
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr8_+_19796730
|
8.155
|
|
LPL
|
lipoprotein lipase
|
|
chr10_-_14050453
|
8.143
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr21_-_48024985
|
8.142
|
NM_006272
|
S100B
|
S100 calcium binding protein B
|
|
chr3_+_123813527
|
7.995
|
NM_001024660
NM_003947
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr14_-_22005035
|
7.913
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr1_+_204913353
|
7.860
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr7_-_135194823
|
7.824
|
NM_001008225
NM_001190847
NM_001190848
NM_001190849
NM_001190850
NM_013316
|
CNOT4
|
CCR4-NOT transcription complex, subunit 4
|
|
chr1_-_156399183
|
7.818
|
NM_006365
|
C1orf61
|
chromosome 1 open reading frame 61
|
|
chr4_-_100273831
|
7.738
|
NM_000669
|
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide
|
|
chr6_-_13408368
|
7.700
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr19_-_51531283
|
7.690
|
NM_001167605
NM_006853
|
KLK11
|
kallikrein-related peptidase 11
|
|
chr4_+_88754120
|
7.664
|
NM_001184695
NM_001184696
NM_001184697
NM_020203
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr6_-_31514313
|
7.565
|
|
ATP6V1G2-DDX39B
ATP6V1G2
|
ATP6V1G2-DDX39B readthrough (non-protein coding)
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr3_+_186330844
|
7.519
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr14_-_23652818
|
7.472
|
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr5_+_161494647
|
7.457
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr3_-_139258531
|
7.326
|
NM_001130992
NM_001130993
NM_002899
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr11_+_112832089
|
7.277
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr22_+_31742739
|
7.277
|
|
FLJ20464
|
uncharacterized protein FLJ20464
|
|
chrX_+_103031753
|
7.243
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr5_+_130506635
|
7.229
|
NM_181705
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chr1_-_230513390
|
7.215
|
NM_024554
|
PGBD5
|
piggyBac transposable element derived 5
|
|
chr3_-_139258477
|
7.205
|
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr5_-_88178964
|
7.198
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr12_-_45270071
|
7.166
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr10_+_7745325
|
7.166
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr8_+_19796581
|
7.117
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr6_-_52710892
|
7.014
|
NM_153699
|
GSTA5
|
glutathione S-transferase alpha 5
|
|
chr12_+_48499655
|
7.013
|
NM_001166686
|
PFKM
|
phosphofructokinase, muscle
|
|
chr6_-_154651214
|
6.971
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr3_+_186330859
|
6.932
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr8_-_40755328
|
6.862
|
NM_001135731
NM_024645
|
ZMAT4
|
zinc finger, matrin-type 4
|
|
chr3_-_52479042
|
6.860
|
NM_020163
|
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G
|
|
chr2_+_162272892
|
6.857
|
|
TBR1
|
T-box, brain, 1
|
|
chr7_-_150946031
|
6.790
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr3_+_38347426
|
6.777
|
NM_004803
|
SLC22A14
|
solute carrier family 22, member 14
|
|
chr6_-_27858569
|
6.774
|
NM_003535
|
HIST1H3J
|
histone cluster 1, H3j
|
|
chr3_+_159557649
|
6.757
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr7_-_137028370
|
6.716
|
|
PTN
|
pleiotrophin
|
|
chr8_-_22550708
|
6.709
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr3_-_12200850
|
6.685
|
NM_003256
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr14_-_23652840
|
6.675
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr14_-_23479328
|
6.617
|
NM_001130706
NM_001130708
NM_021944
|
C14orf93
|
chromosome 14 open reading frame 93
|
|
chr12_-_45269340
|
6.586
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr14_+_29236171
|
6.558
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr10_+_111969988
|
6.538
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr20_-_52687014
|
6.468
|
|
BCAS1
|
breast carcinoma amplified sequence 1
|
|
chr15_+_69222838
|
6.462
|
NM_145658
NM_001184780
|
SPESP1
NOX5
|
sperm equatorial segment protein 1
NADPH oxidase, EF-hand calcium binding domain 5
|
|
chr5_-_95768660
|
6.393
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr17_-_4693595
|
6.383
|
|
LOC284014
|
uncharacterized LOC284014
|
|
chr11_+_113930430
|
6.379
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chrX_+_110366304
|
6.355
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr3_-_114478117
|
6.343
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_-_146435589
|
6.324
|
NM_181674
NM_181676
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr5_+_54455945
|
6.320
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr9_+_137967088
|
6.296
|
NM_014279
NM_006334
|
OLFM1
|
olfactomedin 1
|
|
chr1_+_11866240
|
6.293
|
|
CLCN6
|
chloride channel 6
|
|
chr3_+_115342302
|
6.291
|
|
GAP43
|
growth associated protein 43
|
|
chr18_-_24445652
|
6.283
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr2_-_224467142
|
6.244
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr4_+_106473776
|
6.174
|
NM_001242729
NM_017700
|
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38
|
|
chr2_-_145274916
|
6.168
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr15_+_78632625
|
6.157
|
NM_004378
|
CRABP1
|
cellular retinoic acid binding protein 1
|
|
chr10_-_104178569
|
6.136
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr6_-_116601102
|
6.129
|
NM_003309
|
TSPYL1
|
TSPY-like 1
|
|
chr7_-_150945731
|
6.043
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr11_+_131240370
|
6.042
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr6_+_31982538
|
6.027
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr17_+_41857802
|
6.027
|
NM_001136483
|
C17orf105
|
chromosome 17 open reading frame 105
|
|
chr4_+_55095263
|
6.015
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr17_-_1619473
|
5.987
|
|
MIR22HG
|
MIR22 host gene (non-protein coding)
|
|
chrX_-_32430370
|
5.929
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr4_-_155511838
|
5.905
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr1_+_65730423
|
5.874
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr6_-_166796363
|
5.851
|
|
BRP44L
|
brain protein 44-like
|
|
chr10_-_62149487
|
5.848
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_+_28317687
|
5.802
|
NM_001242894
NM_001242895
NM_024493
|
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3
|
|
chr1_+_97187174
|
5.798
|
NM_021190
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr16_+_24267474
|
5.793
|
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr9_+_2717525
|
5.774
|
NM_133497
|
KCNV2
|
potassium channel, subfamily V, member 2
|
|
chr5_+_140797270
|
5.771
|
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|