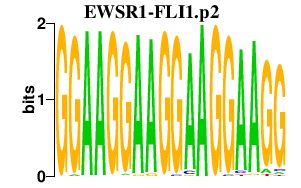
|
chr11_+_73357222
|
49.908
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr3_-_42307661
|
34.506
|
NM_000729
|
CCK
|
cholecystokinin
|
|
chr8_+_104152876
|
18.693
|
NM_001024372
NM_024812
|
BAALC
|
brain and acute leukemia, cytoplasmic
|
|
chr2_+_65215577
|
13.532
|
NM_001193493
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr2_+_79740059
|
13.261
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr3_+_115342302
|
11.639
|
|
GAP43
|
growth associated protein 43
|
|
chr19_-_47551881
|
10.982
|
NM_017854
|
TMEM160
|
transmembrane protein 160
|
|
chr12_-_49351297
|
9.353
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr12_-_49351129
|
9.047
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr12_-_49351263
|
8.955
|
|
|
|
|
chr12_-_49351251
|
8.588
|
NM_001659
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr18_-_5630629
|
8.509
|
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr12_-_49351240
|
8.354
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr7_+_16793320
|
8.277
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chrX_-_148586666
|
8.269
|
NM_000202
NM_001166550
NM_006123
|
IDS
|
iduronate 2-sulfatase
|
|
chr1_-_156470565
|
8.035
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr12_-_49351245
|
7.947
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr1_-_20812727
|
7.918
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr6_-_134496833
|
7.896
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_-_16759939
|
7.883
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_+_37571752
|
7.878
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr5_-_142000899
|
7.840
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr4_+_166299942
|
7.720
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr17_+_2497233
|
7.628
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr6_-_134497069
|
7.618
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr10_+_23384377
|
6.715
|
NM_012228
|
MSRB2
|
methionine sulfoxide reductase B2
|
|
chr19_-_51472816
|
6.316
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr1_-_68299047
|
6.234
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr20_+_10199455
|
6.170
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr7_+_24323806
|
6.121
|
NM_000905
|
NPY
|
neuropeptide Y
|
|
chr1_-_68697997
|
6.111
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chrX_+_56589961
|
5.949
|
NM_013444
|
UBQLN2
|
ubiquilin 2
|
|
chr11_-_62474810
|
5.669
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr4_+_154125589
|
5.552
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr11_-_62474666
|
5.260
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr5_+_86563594
|
5.239
|
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr2_+_79740171
|
5.205
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr1_+_44412418
|
5.113
|
NM_014652
|
IPO13
|
importin 13
|
|
chr17_-_8063946
|
4.842
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr2_+_182756665
|
4.770
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr15_+_84115979
|
4.765
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr11_+_62475165
|
4.749
|
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr10_+_88718287
|
4.702
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr3_-_56502364
|
4.459
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr1_+_223889253
|
4.429
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr11_-_62474869
|
4.283
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr14_+_29236882
|
4.173
|
|
FOXG1
|
forkhead box G1
|
|
chr11_-_31832592
|
4.152
|
|
PAX6
|
paired box 6
|
|
chr10_+_106014467
|
4.028
|
NM_001191002
NM_004832
|
GSTO1
|
glutathione S-transferase omega 1
|
|
chr4_-_16900028
|
4.027
|
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_57522469
|
3.712
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr1_+_19972259
|
3.700
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr11_-_62475060
|
3.654
|
NM_001122955
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr14_+_23775970
|
3.646
|
NM_001199864
NM_004050
NM_001199839
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough
BCL2-like 2
|
|
chr12_+_16035495
|
3.602
|
|
STRAP
|
serine/threonine kinase receptor associated protein
|
|
chr18_+_12947972
|
3.566
|
NM_001013437
NM_031216
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr10_+_70883930
|
3.455
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr11_-_31832758
|
3.367
|
|
PAX6
|
paired box 6
|
|
chr2_+_182756468
|
3.362
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chr4_-_16900348
|
3.356
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr6_-_143266283
|
3.355
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chrX_+_67718868
|
3.294
|
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr5_+_65222381
|
3.237
|
NM_001006600
NM_001253697
NM_001253698
NM_001253699
NM_001253701
NM_018695
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr20_-_47804675
|
3.192
|
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chrX_-_62974987
|
3.174
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr16_-_70719854
|
3.098
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr3_-_192445344
|
3.031
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr10_+_70883977
|
3.020
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr10_+_70883987
|
2.998
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr10_+_70883856
|
2.997
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr1_-_26232890
|
2.946
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr1_-_226926875
|
2.928
|
NM_002221
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr3_+_33155559
|
2.902
|
|
CRTAP
|
cartilage associated protein
|
|
chr19_-_291168
|
2.897
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr3_-_164914448
|
2.869
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr6_-_32143899
|
2.843
|
NM_006411
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr20_-_47804903
|
2.839
|
NM_001037328
NM_004602
NM_017452
NM_017453
NM_017454
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr6_-_32143886
|
2.823
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr18_+_12948010
|
2.781
|
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr3_-_119396220
|
2.743
|
NM_005694
|
COX17
|
COX17 cytochrome c oxidase assembly homolog (S. cerevisiae)
|
|
chr15_+_41523465
|
2.698
|
|
CHP
|
calcium binding protein P22
|
|
chr13_+_29233279
|
2.681
|
|
POMP
|
proteasome maturation protein
|
|
chr11_+_62475113
|
2.664
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr16_-_47007567
|
2.603
|
NM_005880
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr12_+_49658865
|
2.601
|
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr17_+_48133357
|
2.476
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr8_-_22550057
|
2.457
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr15_+_41523401
|
2.450
|
NM_007236
|
CHP
|
calcium binding protein P22
|
|
chr12_-_49177876
|
2.423
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chr10_+_70883943
|
2.399
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr11_-_77122877
|
2.366
|
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr3_-_100120224
|
2.348
|
NM_014820
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae)
|
|
chr17_+_48133331
|
2.295
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr4_-_16900256
|
2.254
|
|
LDB2
|
LIM domain binding 2
|
|
chr1_-_225840660
|
2.210
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr6_-_32143795
|
2.104
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr7_+_69064316
|
2.093
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr3_-_48471401
|
2.091
|
NM_001130082
|
PLXNB1
|
plexin B1
|
|
chr4_-_85887543
|
2.090
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr13_+_114145307
|
2.072
|
NM_017905
|
TMCO3
|
transmembrane and coiled-coil domains 3
|
|
chr6_-_10412606
|
2.031
|
NM_001032280
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr12_+_2904101
|
1.978
|
NM_002014
|
FKBP4
|
FK506 binding protein 4, 59kDa
|
|
chr9_+_34179036
|
1.937
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr19_-_291335
|
1.933
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr1_-_157015161
|
1.929
|
NM_014784
NM_198236
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11
|
|
chr12_+_49658862
|
1.903
|
NM_032704
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr14_+_32546445
|
1.901
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr3_-_192126837
|
1.896
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr17_+_38137081
|
1.887
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr13_+_29233140
|
1.869
|
NM_015932
|
POMP
|
proteasome maturation protein
|
|
chr5_+_86564044
|
1.831
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr17_+_38137053
|
1.822
|
NM_002809
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr1_+_110168048
|
1.819
|
NM_203404
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr17_+_38137088
|
1.800
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr17_-_56595192
|
1.766
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr17_+_38137106
|
1.690
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr17_+_4843883
|
1.675
|
|
RNF167
|
ring finger protein 167
|
|
chrX_-_62975005
|
1.672
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr17_+_38137115
|
1.659
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr3_-_15900928
|
1.650
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr15_+_59359061
|
1.639
|
|
RNF111
|
ring finger protein 111
|
|
chr6_-_32143676
|
1.618
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr17_+_57697264
|
1.617
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr15_+_84116172
|
1.611
|
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr11_-_64410785
|
1.586
|
NM_138734
|
NRXN2
|
neurexin 2
|
|
chr1_-_163113129
|
1.512
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_-_26232643
|
1.479
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chrX_+_1401570
|
1.472
|
NM_001161530
NM_172247
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chrX_+_67718623
|
1.436
|
NM_001195214
NM_173834
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr17_+_57697283
|
1.433
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chrX_+_107683015
|
1.419
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr14_-_64009971
|
1.414
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr12_+_123237302
|
1.403
|
NM_003677
|
DENR
|
density-regulated protein
|
|
chr11_+_17757494
|
1.399
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr17_+_2496902
|
1.392
|
NM_000430
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr14_-_64010087
|
1.361
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr3_+_33155447
|
1.361
|
NM_006371
|
CRTAP
|
cartilage associated protein
|
|
chrY_+_1351570
|
1.354
|
NM_001161530
NM_172247
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr7_+_69063768
|
1.320
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr1_-_152009490
|
1.317
|
NM_005620
|
S100A11
|
S100 calcium binding protein A11
|
|
chr20_+_30193083
|
1.316
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr12_+_85673884
|
1.295
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr19_-_52900966
|
1.263
|
|
LOC284373
|
uncharacterized LOC284373
|
|
chr15_+_68908878
|
1.260
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr8_-_29206321
|
1.248
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr9_+_34179023
|
1.236
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr8_-_110986923
|
1.230
|
NM_014379
|
KCNV1
|
potassium channel, subfamily V, member 1
|
|
chr3_-_66024508
|
1.190
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr16_+_24267672
|
1.165
|
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr9_+_34179058
|
1.114
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr1_-_233431458
|
1.096
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr1_+_246887377
|
1.059
|
NM_016002
|
SCCPDH
|
saccharopine dehydrogenase (putative)
|
|
chr19_+_48774653
|
1.050
|
NM_153608
|
ZNF114
|
zinc finger protein 114
|
|
chr9_+_34179002
|
1.029
|
NM_001171201
NM_001171202
NM_001171203
NM_001171204
NM_016525
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr1_+_246887591
|
0.995
|
|
SCCPDH
|
saccharopine dehydrogenase (putative)
|
|
chr1_+_29063443
|
0.987
|
NM_001172828
|
YTHDF2
|
YTH domain family, member 2
|
|
chr7_+_94285620
|
0.959
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr18_+_46065367
|
0.957
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr9_+_35538628
|
0.923
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr20_-_44485953
|
0.920
|
NM_005469
|
ACOT8
|
acyl-CoA thioesterase 8
|
|
chr12_+_93771607
|
0.920
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr19_-_48823331
|
0.908
|
NM_144577
|
CCDC114
|
coiled-coil domain containing 114
|
|
chr12_+_2904289
|
0.901
|
|
FKBP4
|
FK506 binding protein 4, 59kDa
|
|
chr7_+_94285736
|
0.883
|
|
PEG10
|
paternally expressed 10
|
|
chr13_+_88324777
|
0.870
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr16_-_2318412
|
0.854
|
NM_080594
|
RNPS1
|
RNA binding protein S1, serine-rich domain
|
|
chr1_-_152009440
|
0.830
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr12_-_25403809
|
0.817
|
|
KRAS
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
|
|
chr1_-_152009467
|
0.816
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr7_-_150973819
|
0.807
|
NM_001003802
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr17_-_56591900
|
0.799
|
|
MTMR4
|
myotubularin related protein 4
|
|
chr10_+_114710785
|
0.759
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr1_-_223537400
|
0.752
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr1_+_186798031
|
0.727
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr11_-_8832881
|
0.718
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr7_-_107204705
|
0.680
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr9_-_124990706
|
0.672
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr5_+_152870083
|
0.660
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr4_-_74124305
|
0.659
|
|
|
|
|
chr12_+_49209300
|
0.653
|
NM_001206916
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr17_+_30264054
|
0.649
|
|
SUZ12
|
suppressor of zeste 12 homolog (Drosophila)
|
|
chr9_-_130639939
|
0.644
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr17_+_53828339
|
0.636
|
NM_021213
|
PCTP
|
phosphatidylcholine transfer protein
|
|
chr17_+_30264261
|
0.625
|
|
|
|
|
chr16_-_47007483
|
0.613
|
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr3_-_100119705
|
0.612
|
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae)
|
|
chr5_-_137090037
|
0.591
|
NM_006805
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr11_+_47600561
|
0.590
|
NM_004551
|
NDUFS3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase)
|
|
chr2_+_242127923
|
0.582
|
NM_001001666
NM_001001891
|
ANO7
|
anoctamin 7
|
|
chr10_-_126849556
|
0.563
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr3_+_130612806
|
0.557
|
NM_001199179
NM_001001485
NM_001199183
NM_001199184
NM_001199185
NM_014382
NM_001001486
NM_001001487
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr11_+_68857483
|
0.547
|
|
|
|
|
chr1_-_17380486
|
0.507
|
NM_003000
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip)
|
|
chr1_-_152009406
|
0.478
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr18_+_56530677
|
0.469
|
|
ZNF532
|
zinc finger protein 532
|
|
chr12_-_49365545
|
0.456
|
NM_003394
|
WNT10B
|
wingless-type MMTV integration site family, member 10B
|
|
chr16_+_69458489
|
0.437
|
NM_030579
|
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane)
|
|
chr4_-_74124456
|
0.432
|
NM_032217
NM_198889
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr21_+_40177847
|
0.426
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr16_-_2318378
|
0.407
|
|
RNPS1
|
RNA binding protein S1, serine-rich domain
|