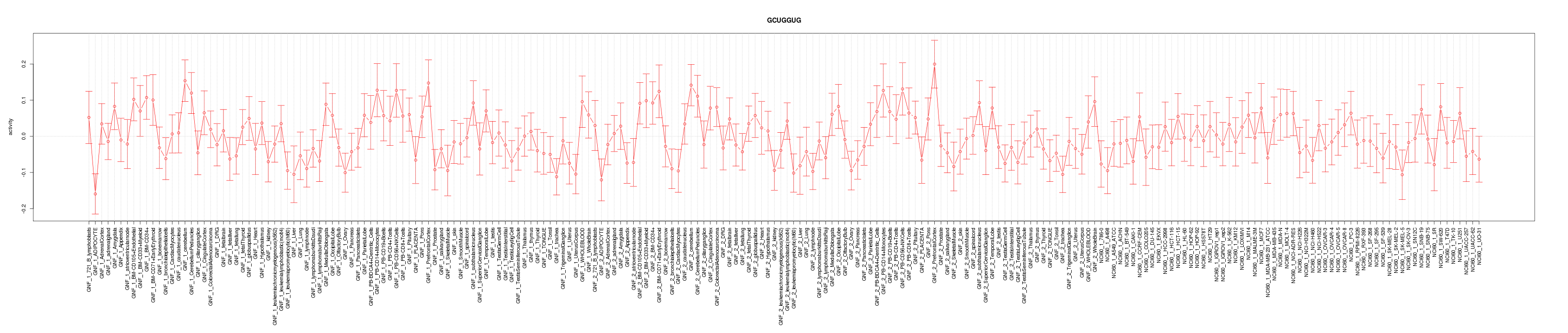
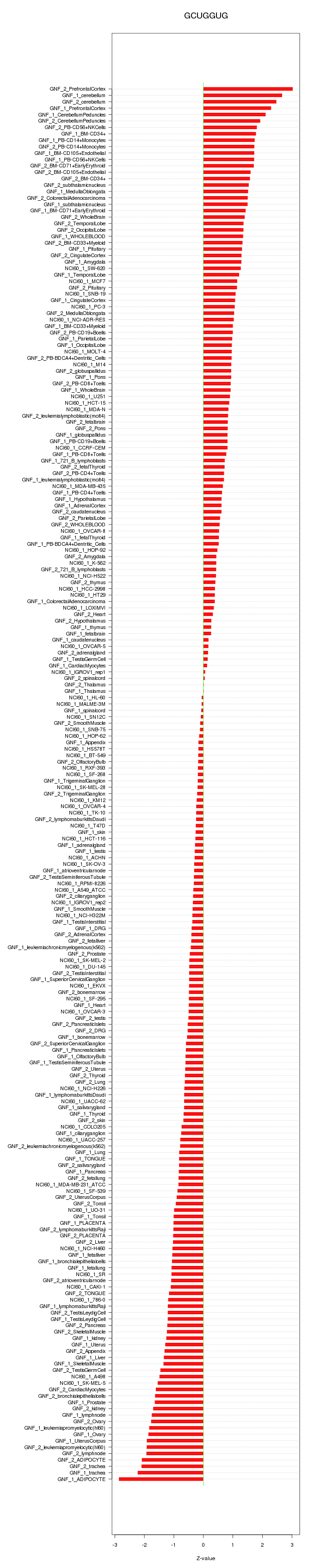
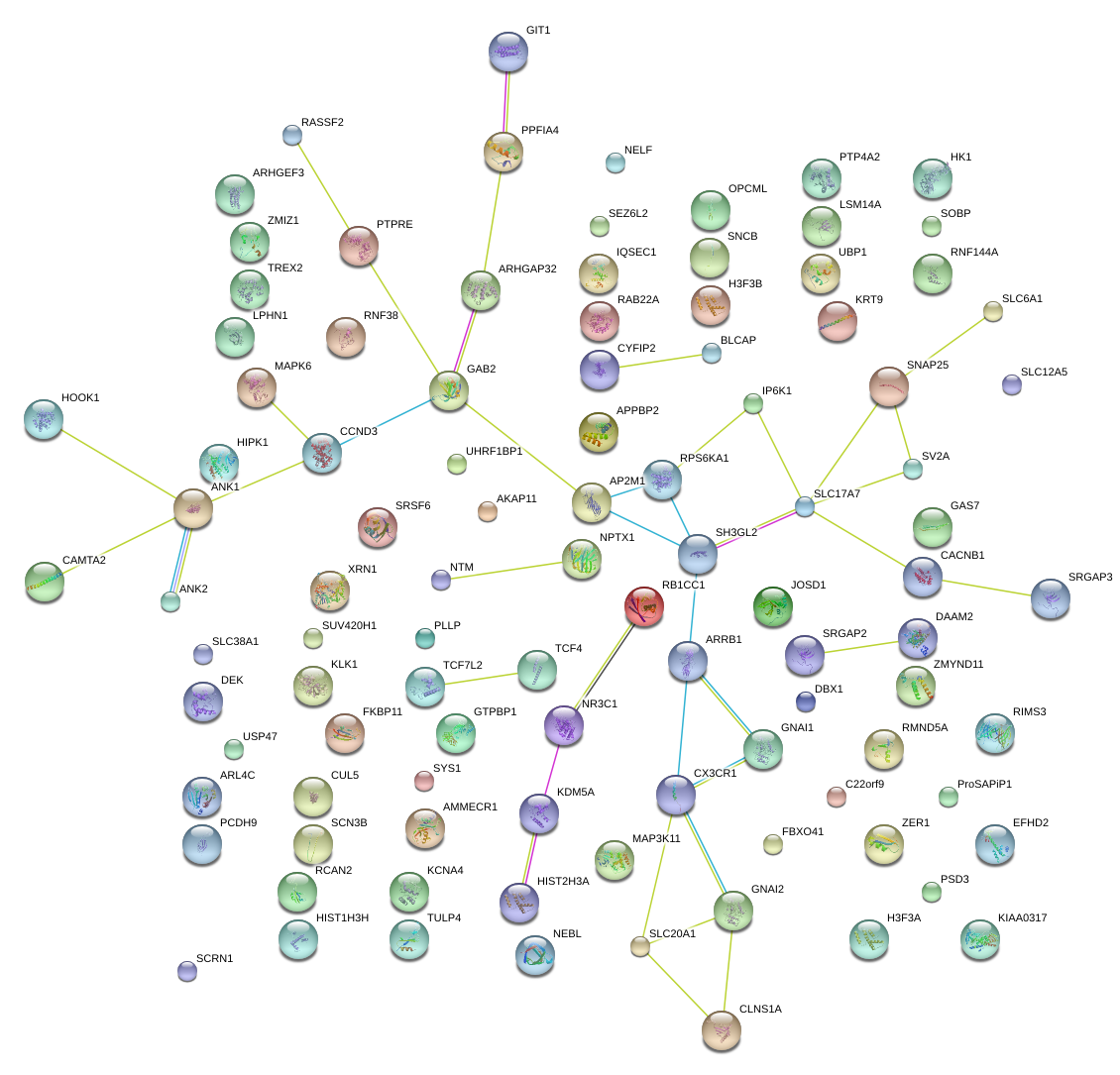
|
chr7_-_30029904
|
16.571
|
NM_001145514
|
SCRN1
|
secernin 1
|
|
chr7_-_30029284
|
15.971
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr20_+_10199455
|
15.829
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr6_-_46293628
|
12.538
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr19_-_49944613
|
11.807
|
NM_020309
|
SLC17A7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7
|
|
chr3_-_13008959
|
11.508
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr1_-_149889362
|
11.447
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr3_-_13114547
|
11.228
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr5_-_176057324
|
9.968
|
NM_001001502
NM_003085
|
SNCB
|
synuclein, beta
|
|
chr17_-_78450247
|
9.340
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr9_-_140353785
|
9.076
|
NM_001130969
NM_001130970
NM_001130971
NM_001178064
NM_015537
|
NELF
|
nasal embryonic LHRH factor
|
|
chr5_+_156696351
|
8.701
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr6_+_39760772
|
8.658
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr22_-_45608346
|
8.552
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr9_+_17578952
|
8.518
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr17_-_9929567
|
8.434
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr5_+_156693051
|
8.306
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr20_-_36152952
|
8.139
|
NM_001167823
|
BLCAP
|
bladder cancer associated protein
|
|
chr10_+_71075609
|
8.071
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr10_+_71078575
|
7.899
|
NM_000188
|
HK1
|
hexokinase 1
|
|
chr22_-_39095991
|
7.862
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr20_-_36156282
|
7.684
|
NM_001167820
NM_006698
|
BLCAP
|
bladder cancer associated protein
|
|
chr6_+_107811272
|
7.673
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr10_+_71029755
|
7.574
|
NM_033497
NM_033498
NM_033500
|
HK1
|
hexokinase 1
|
|
chr6_+_158733691
|
7.514
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr20_-_36156082
|
7.480
|
NM_001167822
NM_001167821
|
BLCAP
|
bladder cancer associated protein
|
|
chr20_-_4795768
|
7.277
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr19_-_14316980
|
7.076
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr6_+_39760148
|
6.968
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr1_-_32385258
|
6.782
|
NM_001195100
NM_001195101
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr1_-_32403975
|
6.781
|
NM_080391
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr2_+_86947387
|
6.538
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr3_-_56809594
|
6.493
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr17_-_9862765
|
6.428
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr3_-_9291251
|
6.377
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr12_-_46663207
|
6.371
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr20_-_4804067
|
6.332
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr1_-_41131323
|
6.247
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr11_-_123525300
|
6.186
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr11_-_75062650
|
6.138
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chr22_-_45636584
|
6.040
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr17_-_9940004
|
5.893
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr17_-_10101854
|
5.846
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr3_-_56835829
|
5.835
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr16_-_57318574
|
5.797
|
NM_015993
|
PLLP
|
plasmolipin
|
|
chr3_-_57113292
|
5.732
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr2_+_113403433
|
5.728
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr13_+_42846255
|
5.579
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr3_+_11034419
|
5.541
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr3_-_33481877
|
5.462
|
NM_001128161
|
UBP1
|
upstream binding protein 1 (LBP-1a)
|
|
chr2_-_235405692
|
5.202
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr17_-_4890920
|
5.202
|
NM_001171166
NM_015099
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr16_-_29910340
|
5.147
|
NM_001114099
NM_001114100
NM_001243332
NM_001243333
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr3_-_33481862
|
5.064
|
NM_001128160
NM_014517
|
UBP1
|
upstream binding protein 1 (LBP-1a)
|
|
chr10_+_129845806
|
5.043
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr3_+_50273645
|
4.964
|
NM_002070
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr20_+_44650260
|
4.946
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr6_-_41909386
|
4.916
|
NM_001136125
NM_001760
|
CCND3
|
cyclin D3
|
|
chr10_+_80828776
|
4.839
|
NM_020338
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr20_+_43991739
|
4.808
|
NM_001099791
NM_033542
|
SYS1-DBNDD2
SYS1
|
SYS1-DBNDD2 readthrough
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr10_-_21463019
|
4.805
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr20_+_44657812
|
4.764
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr20_-_3149070
|
4.739
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr2_+_7057471
|
4.716
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr3_+_50284325
|
4.712
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr17_-_73775822
|
4.659
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr15_+_52311408
|
4.587
|
NM_002748
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr6_+_34759743
|
4.571
|
NM_017754
|
UHRF1BP1
|
UHRF1 binding protein 1
|
|
chr14_-_75179739
|
4.570
|
NM_001039479
|
KIAA0317
|
KIAA0317
|
|
chr17_-_27916602
|
4.484
|
NM_001085454
NM_014030
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr10_+_129705299
|
4.445
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr17_-_4890664
|
4.333
|
NM_001171167
NM_001171168
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr11_-_78052925
|
4.311
|
NM_012296
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr2_-_73496595
|
4.292
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr6_-_42016609
|
4.225
|
NM_001136017
NM_001136126
|
CCND3
|
cyclin D3
|
|
chr4_+_113970784
|
4.203
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr8_-_53626991
|
4.136
|
NM_001083617
NM_014781
|
RB1CC1
|
RB1-inducible coiled-coil 1
|
|
chr1_+_203020310
|
4.097
|
NM_015053
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4
|
|
chr11_+_11862955
|
4.087
|
NM_017944
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr11_-_128894008
|
4.033
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr9_-_131534186
|
4.018
|
NM_006336
|
ZER1
|
zer-1 homolog (C. elegans)
|
|
chr11_-_65381641
|
3.993
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr11_-_129062092
|
3.858
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr11_-_67980699
|
3.855
|
NM_016028
NM_017635
|
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila)
|
|
chr11_+_107879407
|
3.852
|
NM_003478
|
CUL5
|
cullin 5
|
|
chr8_-_18871163
|
3.849
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr3_-_49823626
|
3.842
|
NM_001242829
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr3_+_183892569
|
3.836
|
NM_001025205
NM_004068
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr11_-_77348758
|
3.728
|
NM_001293
|
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A
|
|
chr8_-_18666295
|
3.721
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr1_+_114496498
|
3.716
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr4_+_113739203
|
3.695
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr10_-_21186530
|
3.626
|
NM_006393
|
NEBL
|
nebulette
|
|
chr1_+_26856240
|
3.512
|
NM_002953
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr19_+_34663311
|
3.464
|
NM_001114093
NM_015578
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr1_+_114493766
|
3.461
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr3_-_39323183
|
3.416
|
NM_001171174
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr3_-_39321479
|
3.401
|
NM_001337
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr3_-_39321929
|
3.375
|
NM_001171172
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr11_-_78128852
|
3.372
|
NM_080491
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr13_-_67804432
|
3.354
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr1_+_15736387
|
3.307
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr3_-_39322763
|
3.301
|
NM_001171171
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr12_-_498428
|
3.294
|
NM_001042603
|
KDM5A
|
lysine (K)-specific demethylase 5A
|
|
chr20_+_42086468
|
3.292
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr18_-_53255734
|
3.291
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr5_-_142815076
|
3.264
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr22_+_39101806
|
3.252
|
NM_004286
|
GTPBP1
|
GTP binding protein 1
|
|
chr10_+_180404
|
3.194
|
NM_006624
NM_001202465
NM_001202466
NM_212479
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chrX_-_109683457
|
3.188
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr10_+_181410
|
3.185
|
NM_001202464
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr11_+_131240370
|
3.164
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr3_-_142166782
|
3.149
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr8_-_41754244
|
3.077
|
NM_001142446
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr9_-_36400212
|
3.050
|
NM_022781
NM_194329
|
RNF38
|
ring finger protein 38
|
|
chr20_+_56884749
|
3.022
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr5_-_142783975
|
3.016
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr17_-_58603492
|
3.005
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr6_-_18264638
|
2.990
|
NM_001134709
NM_003472
|
DEK
|
DEK oncogene
|
|
chr14_-_24037266
|
2.930
|
NM_003917
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr1_+_26872342
|
2.894
|
NM_001006665
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr2_+_24807345
|
2.883
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr8_-_41655139
|
2.835
|
NM_000037
NM_020475
NM_020476
NM_020477
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr17_-_2304217
|
2.826
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr3_-_49823972
|
2.790
|
NM_001006115
NM_153273
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr8_+_23101149
|
2.697
|
NM_152272
|
CHMP7
|
charged multivesicular body protein 7
|
|
chr3_+_69134057
|
2.694
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr10_-_12085022
|
2.677
|
NM_080599
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr6_+_15246299
|
2.674
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr2_-_145277879
|
2.623
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr12_+_112856535
|
2.622
|
NM_002834
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr5_-_142783225
|
2.553
|
NM_000176
NM_001020825
NM_001024094
NM_001204258
NM_001204259
NM_001204260
NM_001204261
NM_001204262
NM_001204263
NM_001204264
NM_001204265
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr2_-_240322620
|
2.551
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr11_-_70935807
|
2.526
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr22_-_43355886
|
2.525
|
NM_001184971
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr4_-_2263706
|
2.518
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr5_+_49961732
|
2.485
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr10_-_12084772
|
2.480
|
NM_015542
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr15_+_80696569
|
2.446
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr5_+_154238196
|
2.421
|
NM_004779
|
CNOT8
|
CCR4-NOT transcription complex, subunit 8
|
|
chr15_+_49170264
|
2.419
|
NM_014335
|
EID1
|
EP300 interacting inhibitor of differentiation 1
|
|
chr13_+_36050885
|
2.416
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr22_-_39240016
|
2.365
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr14_-_21852424
|
2.356
|
NM_007192
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr12_-_54653369
|
2.275
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr20_-_1448336
|
2.238
|
NM_001206736
NM_016143
NM_018839
|
NSFL1C
|
NSFL1 (p97) cofactor (p47)
|
|
chr1_+_39547088
|
2.226
|
NM_012090
|
MACF1
|
microtubule-actin crosslinking factor 1
|
|
chr3_+_113465860
|
2.223
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr8_-_41522724
|
2.213
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr15_-_43877043
|
2.190
|
NM_014659
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1
|
|
chr1_-_20812727
|
2.181
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr1_-_154531083
|
2.179
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr1_+_163038934
|
2.170
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr19_+_18208596
|
2.140
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr22_-_43343046
|
2.130
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chrX_-_118827322
|
2.122
|
NM_015129
NM_145799
NM_145800
NM_145802
|
SEPT6
|
septin 6
|
|
chr1_+_163038564
|
2.088
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_-_120315074
|
2.087
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr5_+_6746100
|
2.086
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr1_+_163041694
|
2.085
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr3_+_140660639
|
2.054
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr1_+_163038395
|
2.015
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr10_-_65225605
|
2.014
|
NM_032776
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr17_+_43138675
|
2.012
|
NM_021079
|
NMT1
|
N-myristoyltransferase 1
|
|
chr5_-_74162614
|
2.004
|
NM_015566
|
FAM169A
|
family with sequence similarity 169, member A
|
|
chr20_+_17206751
|
2.004
|
NM_001201528
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr9_-_36401149
|
1.947
|
NM_194330
|
RNF38
|
ring finger protein 38
|
|
chr5_+_49962771
|
1.938
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr1_+_33116748
|
1.908
|
NM_001135255
NM_005610
NM_001135256
|
RBBP4
|
retinoblastoma binding protein 4
|
|
chr3_-_195635879
|
1.873
|
NM_005781
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr5_+_6714717
|
1.831
|
NM_001171805
NM_006999
|
PAPD7
|
PAP associated domain containing 7
|
|
chr19_+_41768368
|
1.824
|
NM_144732
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr8_+_106331146
|
1.818
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr11_-_46142955
|
1.808
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr17_-_8301143
|
1.793
|
NM_001146684
|
RNF222
|
ring finger protein 222
|
|
chr1_-_1624180
|
1.784
|
NM_001110781
|
SLC35E2B
|
solute carrier family 35, member E2B
|
|
chr19_+_35521591
|
1.775
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr3_+_23987514
|
1.771
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr11_-_70507911
|
1.767
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr2_+_219264477
|
1.719
|
NM_021198
NM_182642
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr1_+_28199053
|
1.715
|
NM_001039477
NM_001105556
NM_004848
|
C1orf38
|
chromosome 1 open reading frame 38
|
|
chr13_+_35516390
|
1.707
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr4_+_159131400
|
1.651
|
NM_018342
|
TMEM144
|
transmembrane protein 144
|
|
chr9_-_111882209
|
1.621
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr17_-_56606710
|
1.610
|
NM_004574
|
SEPT4
|
septin 4
|
|
chr17_-_56618178
|
1.601
|
NM_001198713
|
SEPT4
|
septin 4
|
|
chr2_-_43453737
|
1.600
|
NM_006887
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr5_-_73937248
|
1.586
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr11_-_118966175
|
1.579
|
NM_002105
|
H2AFX
|
H2A histone family, member X
|
|
chr14_+_60712469
|
1.575
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr16_-_69419833
|
1.553
|
NM_005652
|
TERF2
|
telomeric repeat binding factor 2
|
|
chr3_-_195622431
|
1.551
|
NM_001010938
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr12_-_54653317
|
1.547
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr4_-_42658883
|
1.547
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr2_+_219263060
|
1.526
|
NM_001206878
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chrX_-_109561314
|
1.515
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr2_+_74756531
|
1.507
|
NM_013247
NM_145074
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr11_+_118978087
|
1.488
|
NM_014807
|
C2CD2L
|
C2CD2-like
|
|
chr1_-_200992824
|
1.471
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr6_+_42982221
|
1.445
|
NM_001242872
|
KLHDC3
|
kelch domain containing 3
|