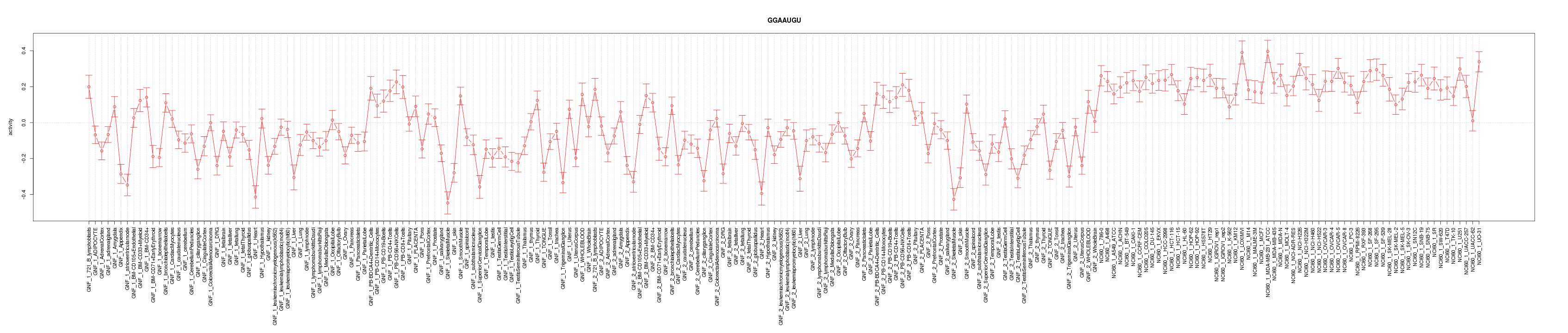
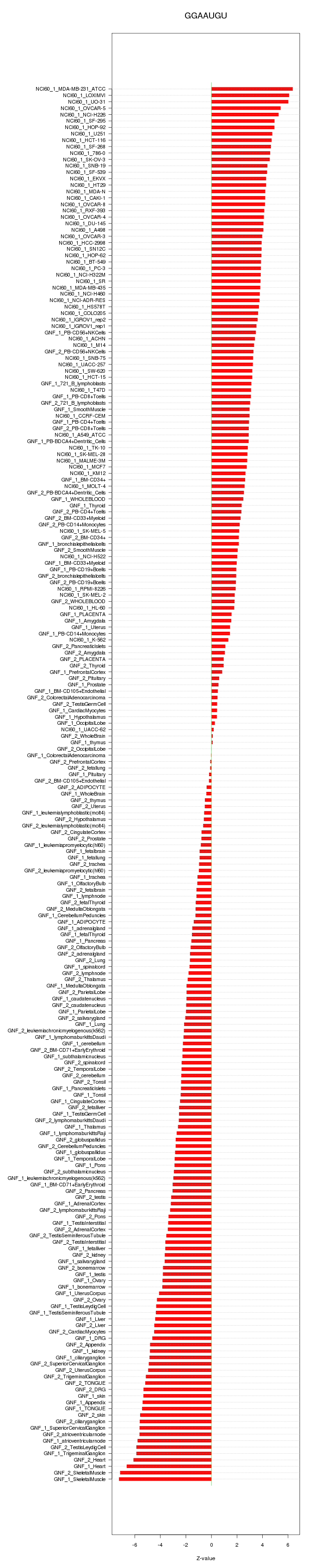
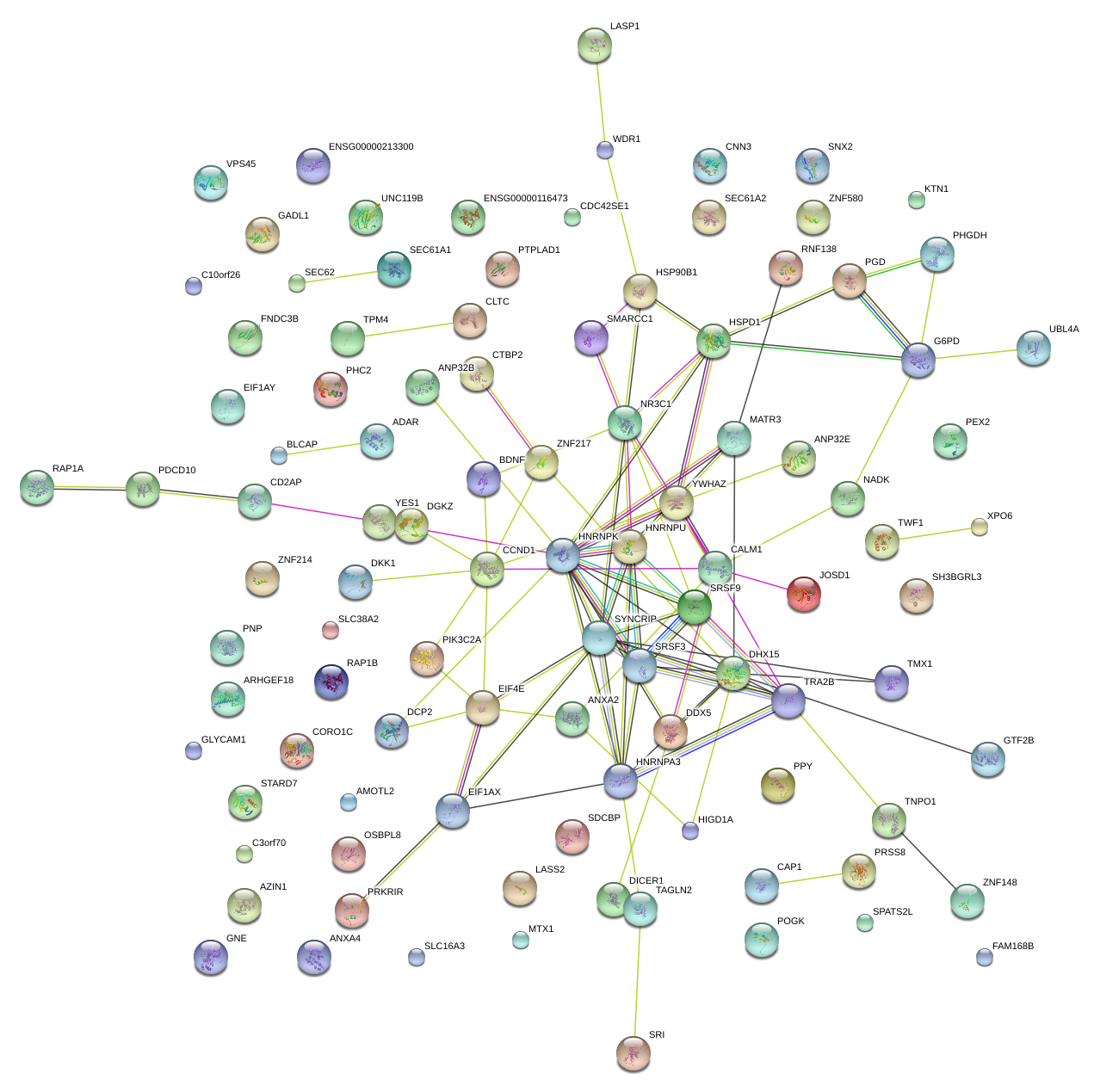
|
chr12_-_109125289
|
101.317
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr8_-_101965601
|
91.768
|
NM_003406
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr6_+_47445440
|
85.981
|
NM_012120
|
CD2AP
|
CD2-associated protein
|
|
chr17_+_37026111
|
84.087
|
NM_006148
|
LASP1
|
LIM and SH3 protein 1
|
|
chr8_-_101965194
|
83.097
|
NM_001135699
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr8_-_101963353
|
81.140
|
NM_001135700
NM_001135701
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr8_-_101962767
|
73.429
|
NM_001135702
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr19_+_16178170
|
68.471
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr3_-_185655833
|
64.393
|
NM_001243879
NM_004593
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr1_-_154578775
|
64.054
|
NM_001193495
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr1_-_150207025
|
63.265
|
NM_001136479
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr1_-_154580677
|
61.901
|
NM_001111
NM_015840
NM_015841
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr5_+_72112417
|
61.357
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr1_+_26606212
|
59.401
|
NM_031286
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr3_+_171757413
|
58.699
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr19_+_16187047
|
58.412
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr1_-_154600446
|
58.142
|
NM_001025107
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr8_-_101964334
|
57.765
|
NM_145690
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr5_+_138629419
|
56.836
|
NM_001194955
NM_001194956
|
MATR3
|
matrin 3
|
|
chr11_+_69455837
|
56.606
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr11_-_17191287
|
55.932
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr5_+_138629330
|
55.813
|
NM_018834
|
MATR3
|
matrin 3
|
|
chr22_-_39095991
|
55.627
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr8_-_103876390
|
54.752
|
NM_015878
NM_148174
|
AZIN1
|
antizyme inhibitor 1
|
|
chrX_-_153775064
|
54.084
|
NM_000402
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr15_+_65822738
|
53.895
|
NM_016395
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1
|
|
chr12_+_104324186
|
53.682
|
NM_003299
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr1_-_150208462
|
53.278
|
NM_001136478
NM_030920
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr18_+_29671817
|
52.435
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr14_+_56046796
|
51.855
|
NM_001079521
NM_001079522
NM_004986
NM_182926
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr12_-_120907535
|
50.902
|
NM_003769
|
SRSF9
|
serine/arginine-rich splicing factor 9
|
|
chr5_+_138609440
|
50.793
|
NM_001194954
NM_199189
|
MATR3
|
matrin 3
|
|
chr5_+_72143929
|
50.668
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr15_-_60690115
|
50.531
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr1_-_151032082
|
50.442
|
NM_001038707
NM_020239
|
CDC42SE1
|
CDC42 small effector 1
|
|
chr1_-_159895279
|
49.929
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chr1_+_10459084
|
49.469
|
NM_002631
|
PGD
|
phosphogluconate dehydrogenase
|
|
chr3_-_184870727
|
48.725
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr2_-_131850988
|
47.480
|
NM_001009993
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr1_-_89357246
|
47.281
|
NM_001514
|
GTF2B
|
general transcription factor IIB
|
|
chr18_+_29672568
|
46.672
|
NM_001191324
NM_198128
|
RNF138
|
ring finger protein 138
|
|
chr19_+_56153417
|
46.478
|
NM_016202
NM_001163423
|
ZNF580
|
zinc finger protein 580
|
|
chr10_-_126849066
|
45.413
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr3_+_169684549
|
45.374
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr14_-_95599794
|
44.987
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr3_+_171758289
|
44.287
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr17_+_57697036
|
43.581
|
NM_004859
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr2_+_69969105
|
43.151
|
NM_001153
|
ANXA4
|
annexin A4
|
|
chr19_+_7504573
|
43.095
|
NM_001130955
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr9_+_100745456
|
42.444
|
NM_006401
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B
|
|
chr1_-_1711453
|
42.275
|
NM_001198994
|
NADK
|
NAD kinase
|
|
chr14_+_51706885
|
42.109
|
NM_030755
|
TMX1
|
thioredoxin-related transmembrane protein 1
|
|
chr2_-_198364997
|
41.953
|
NM_199440
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr9_-_86595485
|
41.145
|
NM_002140
NM_031262
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr1_-_245027808
|
40.621
|
NM_004501
NM_031844
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr1_+_166808721
|
40.238
|
NM_017542
|
POGK
|
pogo transposable element with KRAB domain
|
|
chr3_+_127771126
|
40.072
|
NM_013336
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr3_-_134093235
|
39.937
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr2_-_198364623
|
39.896
|
NM_002156
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr3_-_167452593
|
39.878
|
NM_007217
NM_145860
NM_145859
|
PDCD10
|
programmed cell death 10
|
|
chr14_+_20937537
|
39.532
|
NM_000270
|
PNP
|
purine nucleoside phosphorylase
|
|
chr7_-_87856282
|
38.898
|
NM_198901
|
SRI
|
sorcin
|
|
chr5_+_122110737
|
38.816
|
NM_003100
|
SNX2
|
sorting nexin 2
|
|
chr17_+_80193649
|
38.726
|
NM_001206952
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr17_+_80192774
|
37.945
|
NM_001206951
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chrX_-_153775774
|
37.850
|
NM_001042351
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr1_-_150947456
|
37.755
|
NM_022075
NM_181746
|
CERS2
|
ceramide synthase 2
|
|
chr9_-_36276930
|
37.706
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr10_+_104535887
|
37.371
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr4_-_99851785
|
37.209
|
NM_001130679
NM_001968
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr10_-_126716452
|
37.116
|
NM_022802
|
CTBP2
|
C-terminal binding protein 2
|
|
chr1_-_95392501
|
36.803
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr9_-_86595167
|
36.470
|
NM_031263
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr4_-_24586178
|
36.252
|
NM_001358
|
DHX15
|
DEAH (Asp-Glu-Ala-His) box polypeptide 15
|
|
chr11_+_46383115
|
35.540
|
NM_001105540
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr3_-_42845911
|
35.215
|
NM_001099668
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A
|
|
chr6_+_36562089
|
35.028
|
NM_003017
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr1_+_155178489
|
34.886
|
NM_002455
NM_198883
|
MTX1
|
metaxin 1
|
|
chr2_-_96874569
|
34.584
|
NM_020151
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr5_-_142783975
|
34.232
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr12_+_69004614
|
33.901
|
NM_001010942
NM_001251917
NM_001251918
NM_001251921
NM_001251922
NM_015646
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr17_+_80190107
|
33.868
|
NM_001042423
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr20_-_36152952
|
33.780
|
NM_001167823
|
BLCAP
|
bladder cancer associated protein
|
|
chr17_-_62502410
|
33.649
|
NM_004396
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
|
|
chr3_-_42845946
|
32.994
|
NM_001099669
NM_014056
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A
|
|
chr18_-_812268
|
32.641
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr20_-_52199635
|
32.603
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr19_+_7459964
|
32.440
|
NM_015318
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr9_-_36258383
|
32.427
|
NM_001190383
NM_001190384
NM_005476
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr12_-_76953561
|
32.426
|
NM_001003712
NM_020841
|
OSBPL8
|
oxysterol binding protein-like 8
|
|
chr11_-_27741192
|
32.005
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_+_112312392
|
32.004
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr12_-_46766563
|
31.953
|
NM_018976
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr12_-_44200047
|
31.651
|
NM_001242397
NM_002822
|
TWF1
|
twinfilin, actin-binding protein, homolog 1 (Drosophila)
|
|
chr4_-_10118500
|
31.497
|
NM_005112
NM_017491
|
WDR1
|
WD repeat domain 1
|
|
chr12_+_121147830
|
31.128
|
NM_001080533
|
UNC119B
|
unc-119 homolog B (C. elegans)
|
|
chr10_+_54074037
|
30.512
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr6_-_86351156
|
30.504
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr6_-_86352635
|
30.458
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_150039341
|
30.430
|
NM_007259
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae)
|
|
chr11_-_76091866
|
30.373
|
NM_004705
|
PRKRIR
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor)
|
|
chr16_-_28223161
|
30.340
|
NM_015171
|
XPO6
|
exportin 6
|
|
chr1_-_33841142
|
30.171
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr2_+_178077421
|
30.141
|
NM_194247
|
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3
|
|
chr3_-_47823386
|
29.842
|
NM_003074
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr14_+_90863302
|
29.792
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr6_-_86352564
|
29.506
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr3_-_125094003
|
29.391
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr8_-_77913279
|
29.367
|
NM_001172086
NM_001172087
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr17_+_80186886
|
29.122
|
NM_004207
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr1_+_40506254
|
29.116
|
NM_001105530
NM_006367
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast)
|
|
chr2_+_201170603
|
29.108
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr2_+_201170795
|
28.908
|
NM_001100422
NM_001100424
NM_001100423
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr8_+_59465727
|
28.564
|
NM_001007067
NM_001007068
NM_001007069
NM_001007070
NM_005625
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr11_-_27681195
|
28.526
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chrX_-_20159757
|
28.510
|
NM_001412
|
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked
|
|
chr10_-_126849556
|
28.187
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr11_-_27721179
|
28.167
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27743604
|
27.817
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr3_-_30936152
|
27.527
|
NM_207359
|
GADL1
|
glutamate decarboxylase-like 1
|
|
chr11_-_85780899
|
27.501
|
NM_001206947
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr11_-_27742224
|
27.369
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr3_-_195163575
|
27.345
|
NM_012287
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2
|
|
chrX_+_123095555
|
27.195
|
NM_001042750
NM_001042751
NM_006603
|
STAG2
|
stromal antigen 2
|
|
chr10_+_5488521
|
27.087
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr10_-_104953010
|
26.817
|
NM_001134373
NM_012229
|
NT5C2
|
5'-nucleotidase, cytosolic II
|
|
chr11_+_844062
|
26.427
|
NM_001025234
|
TSPAN4
|
tetraspanin 4
|
|
chr12_-_49351251
|
26.369
|
NM_001659
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr7_-_86848818
|
26.312
|
NM_024315
|
C7orf23
|
chromosome 7 open reading frame 23
|
|
chr11_+_46366816
|
26.197
|
NM_201533
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr11_+_844445
|
25.598
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr11_+_118230292
|
25.538
|
NM_001204077
NM_004788
|
UBE4A
|
ubiquitination factor E4A
|
|
chr18_+_3447607
|
25.459
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr6_-_100016517
|
24.951
|
NM_001013399
NM_005190
|
CCNC
|
cyclin C
|
|
chr15_-_64455299
|
24.923
|
NM_000942
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr7_+_28475233
|
24.874
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr4_+_39063851
|
24.767
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr14_-_95623665
|
24.355
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr7_-_93633527
|
24.327
|
NM_005868
|
BET1
|
blocked early in transport 1 homolog (S. cerevisiae)
|
|
chr21_+_38455246
|
24.105
|
NM_003316
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr17_-_56065596
|
23.968
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr6_+_71377471
|
23.950
|
NM_001044305
NM_021940
|
SMAP1
|
small ArfGAP 1
|
|
chr18_+_3453771
|
23.943
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr1_-_27816676
|
23.816
|
NM_001201404
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr3_-_196911001
|
23.804
|
NM_001204387
NM_001204388
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr17_+_80186281
|
23.767
|
NM_001042422
NM_001206950
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr21_+_33031932
|
23.609
|
NM_000454
|
SOD1
|
superoxide dismutase 1, soluble
|
|
chr18_+_3412071
|
23.511
|
NM_174886
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr1_+_112162397
|
23.153
|
NM_001010935
NM_002884
|
RAP1A
|
RAP1A, member of RAS oncogene family
|
|
chr5_-_142815076
|
23.116
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr8_+_20054703
|
23.049
|
NM_001693
|
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2
|
|
chr11_+_34073188
|
22.981
|
NM_005898
NM_203364
|
CAPRIN1
|
cell cycle associated protein 1
|
|
chr15_+_89631380
|
22.686
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr16_+_30772932
|
22.685
|
NM_001207033
NM_001207034
NM_014771
|
RNF40
|
ring finger protein 40
|
|
chr17_+_60556247
|
22.499
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr3_-_100120224
|
22.356
|
NM_014820
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae)
|
|
chr22_+_45560529
|
22.190
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr1_-_78444772
|
22.058
|
NM_003902
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr11_+_118443091
|
21.934
|
NM_001142281
NM_001655
|
ARCN1
|
archain 1
|
|
chr7_-_87849366
|
21.920
|
NM_003130
|
SRI
|
sorcin
|
|
chr6_-_53409897
|
21.735
|
NM_001197115
NM_001498
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr1_-_35658742
|
21.681
|
NM_005066
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr17_-_73775822
|
21.626
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr18_+_3449972
|
21.593
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr17_-_47439326
|
21.496
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr11_-_27723061
|
21.477
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr2_+_99225041
|
21.430
|
NM_014044
|
UNC50
|
unc-50 homolog (C. elegans)
|
|
chr17_+_49242795
|
21.176
|
NM_001018137
|
NME2
|
non-metastatic cells 2, protein (NM23B) expressed in
|
|
chr19_+_797390
|
21.104
|
NM_002819
NM_031990
NM_031991
NM_175847
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr1_-_78148326
|
21.036
|
NM_015534
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr6_-_128841432
|
20.798
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chrX_+_12993224
|
20.588
|
NM_021109
|
TMSB4X
|
thymosin beta 4, X-linked
|
|
chr14_-_95608054
|
20.550
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr2_-_26101301
|
20.503
|
NM_018263
|
ASXL2
|
additional sex combs like 2 (Drosophila)
|
|
chr2_+_242255270
|
20.404
|
NM_001008492
NM_004404
|
SEPT2
|
septin 2
|
|
chr19_+_56152281
|
20.354
|
NM_207115
|
ZNF580
|
zinc finger protein 580
|
|
chr15_-_73076113
|
20.296
|
NM_031284
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr18_+_9913954
|
20.296
|
NM_003574
NM_194434
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr18_+_3451589
|
20.193
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr19_+_11094799
|
20.070
|
NM_001128845
NM_001128846
NM_001128847
NM_001128848
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr1_-_52456292
|
20.024
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr7_+_28725597
|
19.896
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr18_+_55714457
|
19.744
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_55816546
|
19.618
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr11_-_27721975
|
19.449
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_145096391
|
19.395
|
NM_004892
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr8_+_98787808
|
18.880
|
NM_018407
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr17_-_37607314
|
18.776
|
NM_004774
|
MED1
|
mediator complex subunit 1
|
|
chr2_+_36583369
|
18.603
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr1_-_1822516
|
18.436
|
NM_002074
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1
|
|
chr17_-_53499055
|
18.297
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr11_+_46354454
|
18.051
|
NM_201532
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chrX_+_123094377
|
18.026
|
NM_001042749
|
STAG2
|
stromal antigen 2
|
|
chr11_+_842821
|
17.976
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr11_-_67211291
|
17.841
|
NM_001018070
|
CORO1B
|
coronin, actin binding protein, 1B
|
|
chr12_-_76425375
|
17.732
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr7_-_105925410
|
17.684
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr12_-_118810712
|
17.601
|
NM_016281
|
TAOK3
|
TAO kinase 3
|
|
chr10_+_5454516
|
17.544
|
NM_001047160
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr8_-_12973752
|
17.452
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|