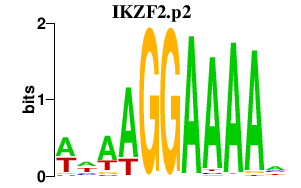
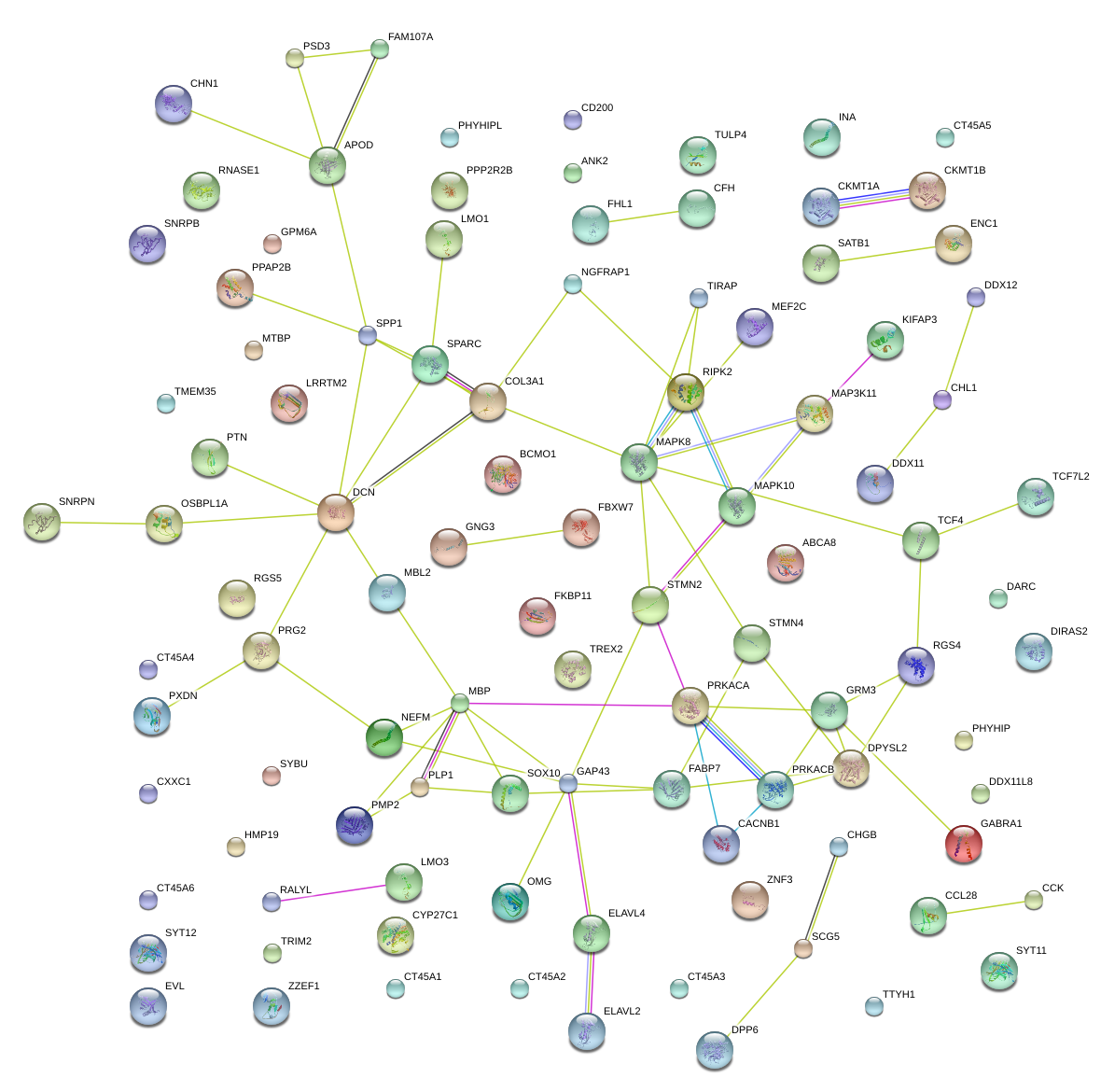
|
chr8_+_80523376
|
34.000
|
|
STMN2
|
stathmin-like 2
|
|
chr11_-_62472901
|
25.514
|
|
|
|
|
chr1_+_155853546
|
24.916
|
|
SYT11
|
synaptotagmin XI
|
|
chr3_-_195310985
|
24.246
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr18_-_74728963
|
22.298
|
|
MBP
|
myelin basic protein
|
|
chr18_-_74729049
|
22.076
|
NM_001025081
NM_001025090
NM_001025092
NM_002385
|
MBP
|
myelin basic protein
|
|
chr4_-_176734181
|
21.662
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr7_+_86273219
|
21.458
|
NM_000840
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr5_-_42811959
|
21.057
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chrX_+_103031433
|
20.696
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chrX_+_103031797
|
20.470
|
|
PLP1
|
proteolipid protein 1
|
|
chr1_-_93399531
|
20.233
|
|
|
|
|
chr3_-_195310770
|
20.078
|
|
APOD
|
apolipoprotein D
|
|
chr3_-_58563070
|
19.754
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chrX_+_103031753
|
19.683
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr4_-_153303663
|
19.424
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chrX_+_73164162
|
18.497
|
|
JPX
|
JPX transcript, XIST activator (non-protein coding)
|
|
chr7_-_64023444
|
17.920
|
|
|
|
|
chr5_-_146460991
|
17.574
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr7_-_137028275
|
17.511
|
|
PTN
|
pleiotrophin
|
|
chr9_-_93405066
|
17.216
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr3_-_58563490
|
16.601
|
NM_001076778
NM_007177
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr4_-_176923572
|
16.300
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr7_-_137028433
|
16.224
|
|
PTN
|
pleiotrophin
|
|
chr17_-_29624249
|
16.211
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr7_-_137028533
|
15.955
|
NM_002825
|
PTN
|
pleiotrophin
|
|
chr11_+_62475165
|
15.697
|
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr2_-_175712276
|
14.899
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr22_-_38380538
|
14.144
|
NM_006941
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chr7_-_137028370
|
14.093
|
|
PTN
|
pleiotrophin
|
|
chr15_+_32933869
|
13.861
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr6_+_158733691
|
13.704
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr1_-_57045235
|
13.489
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr7_+_64498643
|
13.418
|
|
CCT6P3
|
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 3
|
|
chr4_+_88896868
|
13.186
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_-_153273683
|
13.117
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr17_-_66951381
|
12.656
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr8_+_24771276
|
12.500
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr18_-_53089722
|
12.492
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr18_-_53071225
|
12.436
|
NM_001243232
|
TCF4
|
transcription factor 4
|
|
chr15_+_25101697
|
12.187
|
NM_022805
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr1_-_57045214
|
11.812
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_+_161274196
|
11.540
|
NM_000806
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_-_163113938
|
11.484
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr4_+_154125589
|
11.462
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr8_-_18541441
|
11.337
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chrX_+_135251454
|
11.200
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr4_+_88896838
|
10.902
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr7_+_153584383
|
10.851
|
NM_001039350
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr11_+_62475113
|
10.830
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr7_-_99680358
|
10.820
|
|
ZNF3
|
zinc finger protein 3
|
|
chr4_+_113739203
|
10.790
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr5_+_173472692
|
10.754
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr15_+_43885251
|
10.751
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr8_+_85095702
|
10.697
|
|
RALYL
|
RALY RNA binding protein-like
|
|
chr8_+_24771268
|
10.461
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr8_+_80523351
|
10.368
|
|
STMN2
|
stathmin-like 2
|
|
chr4_+_88896899
|
10.261
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr3_-_42305438
|
10.078
|
NM_001174138
|
CCK
|
cholecystokinin
|
|
chr3_+_112051914
|
10.045
|
NM_001004196
NM_005944
|
CD200
|
CD200 molecule
|
|
chr10_+_105036783
|
9.747
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr1_-_170038055
|
9.684
|
NM_001204517
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr8_+_80523048
|
9.669
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr4_-_87281177
|
9.613
|
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr20_+_5891973
|
9.547
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr22_-_38379868
|
9.404
|
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chr11_-_65382701
|
9.266
|
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr16_+_81272293
|
9.229
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr1_+_163038934
|
9.073
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr5_-_151066445
|
9.070
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr1_+_163038564
|
9.046
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_-_16759733
|
9.035
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_+_80523339
|
8.986
|
|
STMN2
|
stathmin-like 2
|
|
chr19_+_54926721
|
8.923
|
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr14_-_21270537
|
8.890
|
NM_198232
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr6_+_123100645
|
8.817
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr1_+_159175048
|
8.816
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr5_-_138211054
|
8.732
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr8_+_121457642
|
8.556
|
NM_022045
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa
|
|
chr4_-_87281374
|
8.498
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr3_+_238274
|
8.458
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr8_+_26435920
|
8.452
|
NM_001244604
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr8_-_82359620
|
8.425
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr12_-_16760935
|
8.412
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr11_+_126152979
|
8.340
|
NM_001039661
NM_148910
|
TIRAP
|
toll-interleukin 1 receptor (TIR) domain containing adaptor protein
|
|
chr2_-_127963342
|
8.318
|
NM_001001665
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1
|
|
chr8_-_22089517
|
8.210
|
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr8_-_110655418
|
8.163
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr1_+_50569581
|
8.147
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr14_+_100531764
|
8.126
|
|
EVL
|
Enah/Vasp-like
|
|
chr3_-_18480203
|
8.078
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chrX_+_100333835
|
8.072
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr12_-_91573264
|
8.065
|
NM_133503
|
DCN
|
decorin
|
|
chr8_-_27101240
|
8.063
|
|
STMN4
|
stathmin-like 4
|
|
chr18_-_21852189
|
8.050
|
NM_018030
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr4_+_88896801
|
8.008
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr3_+_361365
|
7.995
|
NM_001253388
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr14_+_100531745
|
7.915
|
NM_016337
|
EVL
|
Enah/Vasp-like
|
|
chr5_-_43412417
|
7.870
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chrX_+_102632108
|
7.818
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr12_-_16759430
|
7.810
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_+_115342302
|
7.784
|
|
GAP43
|
growth associated protein 43
|
|
chr2_+_189839113
|
7.782
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr5_-_88179278
|
7.774
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr18_-_21017816
|
7.771
|
NM_032933
|
TMEM241
|
transmembrane protein 241
|
|
chrX_+_134866213
|
7.763
|
NM_001017436
NM_152582
|
CT45A3
CT45A4
CT45A2
|
cancer/testis antigen family 45, member A3
cancer/testis antigen family 45, member A4
cancer/testis antigen family 45, member A2
|
|
chr2_-_175869910
|
7.762
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr1_+_84647342
|
7.738
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_+_115342150
|
7.727
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr2_+_17935124
|
7.494
|
NM_182625
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr2_+_189839132
|
7.442
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr19_+_54926604
|
7.395
|
NM_001005367
NM_001201461
NM_020659
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr18_-_53068755
|
7.333
|
|
TCF4
|
transcription factor 4
|
|
chr1_-_230850335
|
7.318
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr12_+_101988746
|
7.251
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr1_+_152850797
|
7.081
|
NM_030663
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein
|
|
chr8_+_1952875
|
7.079
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr8_-_37727937
|
6.992
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chrX_+_110366304
|
6.919
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr9_-_79520878
|
6.917
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr5_+_140247767
|
6.910
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr2_+_189839078
|
6.877
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr18_-_40695483
|
6.823
|
NM_002930
|
RIT2
|
Ras-like without CAAX 2
|
|
chr15_+_43985083
|
6.808
|
NM_001015001
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chr10_+_111765725
|
6.772
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr20_-_23402038
|
6.728
|
NM_022080
|
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta
|
|
chr5_+_173472737
|
6.709
|
|
HMP19
|
HMP19 protein
|
|
chr9_-_91793375
|
6.648
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr18_-_52988916
|
6.643
|
NM_001243234
NM_001243235
|
TCF4
|
transcription factor 4
|
|
chr8_+_11141983
|
6.636
|
NM_015458
|
MTMR9
|
myotubularin related protein 9
|
|
chr6_+_27107060
|
6.586
|
NM_003495
|
HIST2H4B
HIST1H4I
|
histone cluster 2, H4b
histone cluster 1, H4i
|
|
chr12_-_16759531
|
6.545
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr16_+_7382750
|
6.538
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr4_+_71587674
|
6.538
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr13_+_97794050
|
6.482
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr7_+_94024201
|
6.472
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr2_+_207406815
|
6.469
|
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr12_-_16759939
|
6.448
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_108745569
|
6.427
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr13_+_58205943
|
6.399
|
|
PCDH17
|
protocadherin 17
|
|
chrX_+_135251795
|
6.378
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr1_-_57045129
|
6.369
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_-_146435589
|
6.313
|
NM_181674
NM_181676
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr18_-_53303129
|
6.231
|
NM_001243226
|
TCF4
|
transcription factor 4
|
|
chr10_-_79789290
|
6.199
|
NM_007055
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa
|
|
chrX_-_6145887
|
6.179
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr13_+_42846255
|
6.172
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr7_+_138482696
|
6.152
|
NM_001085429
|
TMEM213
|
transmembrane protein 213
|
|
chr4_+_113970784
|
6.133
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr8_+_80523304
|
6.133
|
|
STMN2
|
stathmin-like 2
|
|
chr3_+_10068123
|
6.131
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr19_+_38893828
|
6.131
|
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr2_-_2334887
|
6.106
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr12_+_175872
|
6.096
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr12_-_91576579
|
6.072
|
|
DCN
|
decorin
|
|
chr6_-_46825942
|
6.072
|
|
GPR116
|
G protein-coupled receptor 116
|
|
chr6_+_121756744
|
6.069
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr11_-_62475060
|
6.051
|
NM_001122955
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr5_+_140771482
|
6.039
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chr3_+_112052020
|
6.021
|
|
CD200
|
CD200 molecule
|
|
chrX_+_135230736
|
6.018
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr19_-_12662191
|
5.989
|
NM_144976
|
ZNF564
|
zinc finger protein 564
|
|
chrX_-_63005212
|
5.986
|
NM_001173479
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chrX_+_100805415
|
5.976
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr18_-_21891469
|
5.925
|
NM_001242508
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr7_+_142560512
|
5.923
|
|
EPHB6
|
EPH receptor B6
|
|
chr11_-_33890931
|
5.922
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr12_-_133532847
|
5.912
|
NM_183238
NM_001164715
|
ZNF605
|
zinc finger protein 605
|
|
chr12_-_15038724
|
5.892
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr3_+_101546833
|
5.861
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr7_+_123565908
|
5.858
|
NM_001174045
NM_001174046
|
SPAM1
|
sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding)
|
|
chr1_-_57044980
|
5.825
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr12_+_79439432
|
5.793
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr3_+_10068097
|
5.688
|
NM_001018115
NM_033084
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr16_+_10522724
|
5.688
|
NM_024997
|
ATF7IP2
|
activating transcription factor 7 interacting protein 2
|
|
chr7_-_14880954
|
5.677
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr1_+_50571963
|
5.670
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr11_-_111782447
|
5.641
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr5_-_88199868
|
5.614
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_+_176987148
|
5.600
|
|
WDR17
|
WD repeat domain 17
|
|
chrX_+_133507299
|
5.575
|
NM_001015877
NM_032335
NM_032458
|
PHF6
|
PHD finger protein 6
|
|
chr14_+_23299083
|
5.558
|
NM_178336
NM_180982
NM_181304
NM_181305
NM_181306
NM_181307
|
MRPL52
|
mitochondrial ribosomal protein L52
|
|
chr4_+_106631966
|
5.453
|
NM_001031720
|
GSTCD
|
glutathione S-transferase, C-terminal domain containing
|
|
chr13_-_73356070
|
5.445
|
NM_001128226
NM_014953
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chrX_-_102941582
|
5.430
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr8_-_102803438
|
5.348
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr1_+_168148215
|
5.341
|
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae)
|
|
chr4_-_174320616
|
5.337
|
NM_007281
|
SCRG1
|
stimulator of chondrogenesis 1
|
|
chr8_+_39442086
|
5.326
|
NM_001190956
NM_014237
|
ADAM18
|
ADAM metallopeptidase domain 18
|
|
chr19_+_41509825
|
5.259
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chrX_+_135278912
|
5.225
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_-_238322815
|
5.219
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_-_203320249
|
5.218
|
|
|
|
|
chr12_-_45269340
|
5.217
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr1_+_84630014
|
5.200
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chrX_+_38420690
|
5.194
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr9_+_102677457
|
5.165
|
|
STX17
|
syntaxin 17
|
|
chr2_-_238322770
|
5.150
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr3_-_9290592
|
5.149
|
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr19_+_21264952
|
5.139
|
NM_182515
|
ZNF714
|
zinc finger protein 714
|