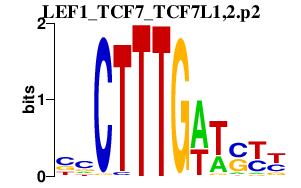
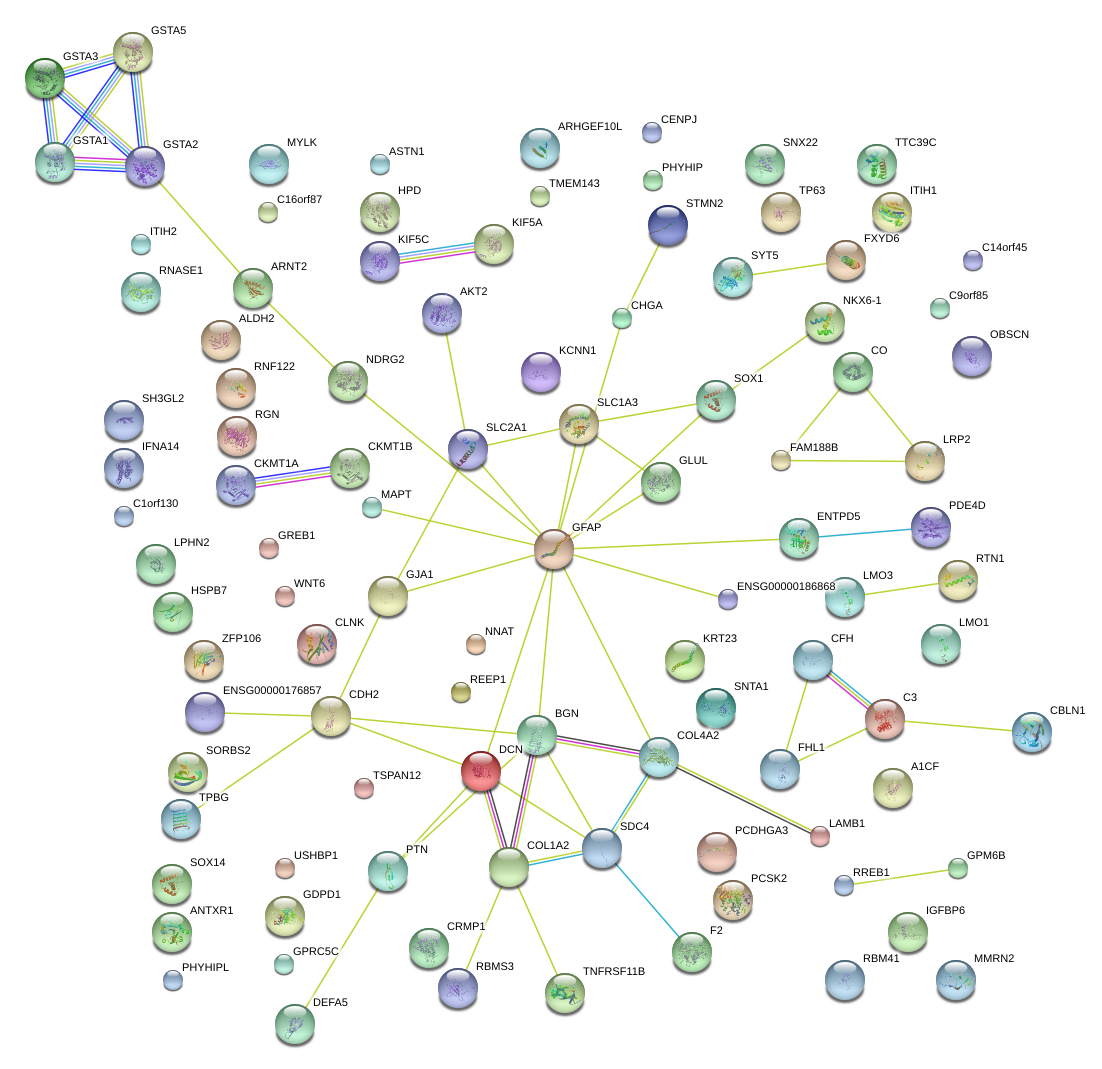
|
chr12_-_91573264
|
22.838
|
NM_133503
|
DCN
|
decorin
|
|
chr9_+_74526527
|
22.391
|
|
C9orf85
|
chromosome 9 open reading frame 85
|
|
chr11_-_117747331
|
20.993
|
|
FXYD6
|
FXYD domain containing ion transport regulator 6
|
|
chr12_-_16759939
|
19.566
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr13_+_111137249
|
19.140
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr17_-_42992852
|
18.401
|
NM_001131019
NM_001242376
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr12_-_16759531
|
18.146
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr18_+_21572736
|
18.123
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr10_+_7745341
|
17.826
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr7_+_94023872
|
17.140
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr4_-_5894695
|
16.129
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr13_+_111134933
|
14.690
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr7_+_94024201
|
14.491
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr1_-_182361311
|
14.089
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chr8_+_80523048
|
13.654
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr10_+_7745354
|
13.588
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr10_+_7745325
|
13.274
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr6_+_7108864
|
13.053
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr12_-_16759430
|
12.971
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_-_22089517
|
12.966
|
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr11_-_76155704
|
12.671
|
|
|
|
|
chr20_+_36149606
|
12.480
|
NM_005386
NM_181689
|
NNAT
|
neuronatin
|
|
chr8_+_80523351
|
12.366
|
|
STMN2
|
stathmin-like 2
|
|
chrX_-_13835012
|
12.359
|
|
GPM6B
|
glycoprotein M6B
|
|
chr5_-_58334936
|
12.239
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr10_+_7745284
|
12.222
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr4_-_186877413
|
12.095
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr8_+_80523304
|
11.998
|
|
STMN2
|
stathmin-like 2
|
|
chr10_+_7745229
|
11.607
|
NM_002216
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr14_-_21271002
|
11.186
|
NM_002933
NM_198234
NM_198235
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr15_-_42783293
|
11.178
|
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr18_-_25757351
|
11.164
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chrX_+_135229536
|
11.142
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr16_-_46865039
|
11.024
|
NM_001001436
|
C16orf87
|
chromosome 16 open reading frame 87
|
|
chr15_+_43885251
|
10.893
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr2_-_86564632
|
10.890
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr10_-_88717319
|
10.713
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr12_-_16759733
|
10.237
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr19_-_17375522
|
10.219
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr8_+_80523339
|
10.161
|
|
STMN2
|
stathmin-like 2
|
|
chr13_-_25496913
|
10.159
|
|
CENPJ
|
centromere protein J
|
|
chr7_-_107643330
|
10.108
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr8_-_119964059
|
10.017
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr12_+_57943843
|
9.877
|
NM_004984
|
KIF5A
|
kinesin family member 5A
|
|
chr5_+_36606667
|
9.717
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr8_-_22089850
|
9.716
|
NM_001099335
NM_014759
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr13_+_111138079
|
9.511
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr2_+_11680079
|
9.414
|
NM_148903
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr19_-_55690717
|
9.398
|
|
SYT5
|
synaptotagmin V
|
|
chr14_+_74486049
|
9.304
|
NM_025057
|
C14orf45
|
chromosome 14 open reading frame 45
|
|
chr6_+_121756744
|
9.292
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr18_-_25756946
|
9.263
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr2_+_69240137
|
9.170
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr1_+_24882565
|
9.125
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr12_+_112204690
|
9.014
|
NM_000690
NM_001204889
|
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial)
|
|
chr14_-_74485813
|
8.913
|
NM_001249
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5
|
|
chr7_-_137028370
|
8.906
|
|
PTN
|
pleiotrophin
|
|
chr7_-_137028533
|
8.869
|
NM_002825
|
PTN
|
pleiotrophin
|
|
chr9_+_17578952
|
8.816
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr17_-_39093671
|
8.637
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr14_-_21270537
|
8.635
|
NM_198232
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr8_-_6914251
|
8.579
|
NM_021010
|
DEFA5
|
defensin, alpha 5, Paneth cell-specific
|
|
chr17_+_44101549
|
8.520
|
|
MAPT
|
microtubule-associated protein tau
|
|
chr7_+_30951414
|
8.400
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr1_-_177133818
|
8.301
|
|
ASTN1
|
astrotactin 1
|
|
chr1_-_177133950
|
8.284
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr17_+_57297827
|
8.279
|
NM_001165993
NM_001165994
NM_182569
|
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1
|
|
chr15_+_80696569
|
8.004
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr1_-_43424324
|
7.988
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr8_-_33424642
|
7.946
|
NM_024787
|
RNF122
|
ring finger protein 122
|
|
chr3_-_123339178
|
7.941
|
|
MYLK
|
myosin light chain kinase
|
|
chr3_-_123339423
|
7.902
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr1_+_82266081
|
7.885
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr19_-_40791145
|
7.861
|
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr9_+_74526369
|
7.859
|
NM_182505
|
C9orf85
|
chromosome 9 open reading frame 85
|
|
chr4_-_10686314
|
7.800
|
NM_052964
|
CLNK
|
cytokine-dependent hematopoietic cell linker
|
|
chr12_+_112204711
|
7.783
|
|
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial)
|
|
chr9_-_21239975
|
7.749
|
NM_002172
|
IFNA14
|
interferon, alpha 14
|
|
chr15_+_64443909
|
7.705
|
NM_024798
|
SNX22
|
sorting nexin 22
|
|
chr5_+_36606456
|
7.701
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr6_+_83073954
|
7.619
|
NM_001166392
|
TPBG
|
trophoblast glycoprotein
|
|
chr2_-_170218972
|
7.505
|
|
LRP2
|
low density lipoprotein receptor-related protein 2
|
|
chr20_-_43977023
|
7.478
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr7_-_137028433
|
7.447
|
|
PTN
|
pleiotrophin
|
|
chr17_+_72428228
|
7.446
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr4_-_85419386
|
7.425
|
NM_006168
|
NKX6-1
|
NK6 homeobox 1
|
|
chr8_+_80523376
|
7.422
|
|
STMN2
|
stathmin-like 2
|
|
chr5_+_140734767
|
7.407
|
NM_018917
NM_032053
|
PCDHGA4
|
protocadherin gamma subfamily A, 4
|
|
chr3_+_189349215
|
7.386
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr1_-_16345284
|
7.316
|
NM_014424
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr8_-_121824015
|
7.294
|
|
|
|
|
chr20_+_17207596
|
7.268
|
NM_001201529
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr14_-_21493122
|
7.194
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr5_+_140771482
|
7.165
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chrX_+_46937744
|
7.157
|
NM_004683
NM_152869
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr12_+_53491417
|
7.095
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr16_-_49315682
|
7.054
|
NM_004352
|
CBLN1
|
cerebellin 1 precursor
|
|
chr19_+_18062110
|
7.031
|
NM_002248
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1
|
|
chr14_+_93389444
|
6.995
|
NM_001275
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr19_-_48867149
|
6.977
|
NM_018273
|
TMEM143
|
transmembrane protein 143
|
|
chr14_-_60097223
|
6.961
|
|
RTN1
|
reticulon 1
|
|
chr3_+_137483133
|
6.913
|
NM_004189
|
SOX14
|
SRY (sex determining region Y)-box 14
|
|
chr19_-_6720585
|
6.811
|
NM_000064
|
C3
|
complement component 3
|
|
chrX_+_152760346
|
6.808
|
NM_001711
|
BGN
|
biglycan
|
|
chr10_-_52645384
|
6.767
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr7_-_120498128
|
6.765
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr3_+_52813936
|
6.761
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr20_-_32031425
|
6.740
|
NM_003098
|
SNTA1
|
syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component)
|
|
chr17_+_7960201
|
6.740
|
|
|
|
|
chr6_-_52668599
|
6.732
|
NM_145740
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chrX_+_152760452
|
6.698
|
|
BGN
|
biglycan
|
|
chr5_-_157002705
|
6.635
|
|
|
|
|
chr1_-_43424536
|
6.608
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr11_+_46740769
|
6.557
|
|
F2
|
coagulation factor II (thrombin)
|
|
chrX_-_106362018
|
6.555
|
|
RBM41
|
RNA binding motif protein 41
|
|
chr1_+_17846818
|
6.555
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr5_+_140753480
|
6.522
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr12_-_16760935
|
6.512
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_+_29322802
|
6.505
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr12_-_122296619
|
6.501
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr2_+_219724545
|
6.494
|
NM_006522
|
WNT6
|
wingless-type MMTV integration site family, member 6
|
|
chr1_-_178062734
|
6.451
|
|
LOC100302401
|
uncharacterized LOC100302401
|
|
chr2_+_169926084
|
6.441
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr3_+_123813527
|
6.416
|
NM_001024660
NM_003947
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr1_+_9299902
|
6.348
|
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase)
|
|
chr13_+_102104941
|
6.327
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr14_-_60097314
|
6.317
|
|
RTN1
|
reticulon 1
|
|
chr11_+_46740736
|
6.277
|
NM_000506
|
F2
|
coagulation factor II (thrombin)
|
|
chr18_-_31802433
|
6.277
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr5_+_140797243
|
6.226
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr6_-_28220001
|
6.219
|
NM_019110
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4
|
|
chrX_-_32173585
|
6.219
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr2_+_16080639
|
6.216
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr17_+_56315786
|
6.204
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr10_-_5045967
|
6.195
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr13_+_39612440
|
6.190
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr11_-_72385411
|
6.190
|
NM_002599
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr19_-_40791265
|
6.105
|
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr7_-_137028275
|
6.064
|
|
PTN
|
pleiotrophin
|
|
chr1_+_93913566
|
6.034
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|
|
chr3_-_167813175
|
6.005
|
NM_014498
|
GOLIM4
|
golgi integral membrane protein 4
|
|
chr1_+_13911966
|
6.000
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr10_-_13749622
|
5.945
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr19_-_40791224
|
5.935
|
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr6_-_24645955
|
5.892
|
NM_001168375
NM_001168376
NM_001168374
NM_001168377
NM_014809
|
KIAA0319
|
KIAA0319
|
|
chr10_+_71561643
|
5.890
|
NM_001130103
NM_080798
NM_080800
NM_080801
NM_080802
NM_080805
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr6_+_41040713
|
5.888
|
|
NFYA
|
nuclear transcription factor Y, alpha
|
|
chr11_+_61129831
|
5.841
|
|
TMEM138
|
transmembrane protein 138
|
|
chr12_+_70759974
|
5.835
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr2_-_128568666
|
5.775
|
|
WDR33
|
WD repeat domain 33
|
|
chr17_+_36861398
|
5.754
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr2_+_149632772
|
5.733
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr15_+_44580870
|
5.688
|
NM_138423
NM_177974
|
CASC4
|
cancer susceptibility candidate 4
|
|
chr11_-_75921785
|
5.656
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr13_-_27334777
|
5.638
|
NM_005288
|
GPR12
|
G protein-coupled receptor 12
|
|
chr1_+_244998967
|
5.631
|
|
FAM36A
|
family with sequence similarity 36, member A
|
|
chr3_-_187388105
|
5.628
|
NM_001048
|
SST
|
somatostatin
|
|
chr10_+_71561687
|
5.602
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr20_+_17206751
|
5.583
|
NM_001201528
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr16_+_72097124
|
5.568
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr1_-_173886401
|
5.543
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr1_+_209878135
|
5.513
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr6_+_27925018
|
5.502
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr8_+_121823538
|
5.499
|
|
|
|
|
chr10_+_14920781
|
5.482
|
NM_001193424
NM_001193426
NM_001193425
NM_001193427
NM_024670
|
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila)
|
|
chr1_-_95392501
|
5.475
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr1_+_244998592
|
5.329
|
NM_198076
|
FAM36A
|
family with sequence similarity 36, member A
|
|
chr19_-_6502224
|
5.325
|
NM_006087
|
TUBB4A
|
tubulin, beta 4A class IVa
|
|
chr6_-_90024936
|
5.316
|
NM_002043
|
GABRR2
|
gamma-aminobutyric acid (GABA) receptor, rho 2
|
|
chr19_-_40791248
|
5.310
|
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr8_+_105352050
|
5.301
|
NM_030788
|
TM7SF4
|
transmembrane 7 superfamily member 4
|
|
chr1_+_150255228
|
5.283
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr2_+_105471745
|
5.271
|
NM_006236
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr12_+_32654976
|
5.257
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr3_+_159557745
|
5.212
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr17_-_2996289
|
5.195
|
NM_002548
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2
|
|
chr1_-_225840660
|
5.149
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr8_-_121823696
|
5.127
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr1_-_173886464
|
5.125
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr1_+_196912910
|
5.111
|
NM_005666
|
CFHR2
|
complement factor H-related 2
|
|
chr20_+_34556528
|
5.097
|
NM_001207076
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr9_-_21217303
|
5.084
|
NM_002173
|
IFNA16
|
interferon, alpha 16
|
|
chr16_-_48419148
|
5.081
|
NM_003031
|
SIAH1
|
seven in absentia homolog 1 (Drosophila)
|
|
chr12_+_62860816
|
5.054
|
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr17_+_77751929
|
5.038
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr8_-_145701717
|
5.031
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chr1_+_19971865
|
5.028
|
NM_001204085
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr2_+_172378756
|
5.012
|
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr19_-_3028901
|
5.006
|
NM_003260
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr11_+_113848221
|
4.972
|
NM_001161772
|
HTR3A
|
5-hydroxytryptamine (serotonin) receptor 3A
|
|
chr12_+_32260084
|
4.941
|
NM_001003398
NM_001714
|
BICD1
|
bicaudal D homolog 1 (Drosophila)
|
|
chr7_-_64451413
|
4.940
|
NM_015852
|
ZNF117
|
zinc finger protein 117
|
|
chrX_+_102632108
|
4.928
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr19_+_37407230
|
4.913
|
NM_001204835
NM_001204836
NM_001204837
NM_001204838
NM_001204839
NM_198539
|
ZNF568
|
zinc finger protein 568
|
|
chr4_-_120243257
|
4.886
|
NM_000134
|
FABP2
|
fatty acid binding protein 2, intestinal
|
|
chr22_-_36013303
|
4.881
|
NM_005368
NM_203378
|
MB
|
myoglobin
|
|
chr5_+_140797270
|
4.857
|
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr17_+_30813928
|
4.827
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr3_+_159557649
|
4.806
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr15_-_89764809
|
4.802
|
NM_000326
|
RLBP1
|
retinaldehyde binding protein 1
|