|
chrX_-_13956445
|
29.669
|
|
|
|
|
chr1_+_1981902
|
23.012
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr16_+_23847271
|
22.292
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chrX_-_13956644
|
22.029
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr16_+_58497628
|
21.817
|
|
NDRG4
|
NDRG family member 4
|
|
chr16_+_58497993
|
21.105
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chrX_-_13956530
|
21.084
|
|
GPM6B
|
glycoprotein M6B
|
|
chr5_+_156693051
|
20.603
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr7_-_766875
|
20.261
|
NM_001164760
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr16_+_58497548
|
20.074
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr19_+_38755238
|
19.512
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_38755097
|
18.908
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr20_+_44034632
|
18.294
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr18_-_74691951
|
18.263
|
|
MBP
|
myelin basic protein
|
|
chr16_+_23847321
|
17.988
|
|
PRKCB
|
protein kinase C, beta
|
|
chr20_+_44035203
|
17.299
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr2_+_79740059
|
16.954
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr19_+_38755422
|
16.764
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr7_-_124405029
|
16.465
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr1_+_65720091
|
16.243
|
|
LOC645195
DNAJC6
|
uncharacterized LOC645195
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr16_-_57318452
|
16.203
|
|
PLLP
|
plasmolipin
|
|
chr16_+_23847359
|
15.749
|
|
PRKCB
|
protein kinase C, beta
|
|
chr2_+_79740171
|
15.496
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr1_+_233749749
|
15.190
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr19_+_54926604
|
14.952
|
NM_001005367
NM_001201461
NM_020659
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chrX_-_118827116
|
14.399
|
|
SEPT6
|
septin 6
|
|
chr4_-_176923572
|
14.042
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr14_+_100150596
|
13.403
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr19_-_46974663
|
13.396
|
|
PNMAL1
|
PNMA-like 1
|
|
chr4_-_36245893
|
13.342
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr14_-_23822025
|
13.073
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr9_-_93405066
|
13.050
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr6_-_3227776
|
13.031
|
NM_178012
|
TUBB2B
|
tubulin, beta 2B class IIb
|
|
chr19_+_47104389
|
12.558
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr9_-_101470836
|
12.347
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr17_+_7554415
|
12.292
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chrX_-_118827155
|
12.200
|
|
SEPT6
|
septin 6
|
|
chr19_-_46974789
|
12.137
|
NM_001103149
NM_018215
|
PNMAL1
|
PNMA-like 1
|
|
chr19_+_47104559
|
12.025
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr7_-_37488894
|
11.942
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_-_767286
|
11.756
|
NM_001164758
NM_001164759
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr4_-_176923480
|
11.694
|
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_176923533
|
11.608
|
|
GPM6A
|
glycoprotein M6A
|
|
chr11_-_694842
|
11.597
|
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr19_-_6502224
|
11.375
|
NM_006087
|
TUBB4A
|
tubulin, beta 4A class IVa
|
|
chr7_-_994008
|
11.328
|
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr4_-_17513676
|
10.844
|
NM_000320
|
QDPR
|
quinoid dihydropteridine reductase
|
|
chr3_+_50712671
|
10.750
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr19_+_47104490
|
10.674
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr5_+_169064238
|
10.647
|
NM_004946
|
DOCK2
|
dedicator of cytokinesis 2
|
|
chr14_-_60097531
|
10.454
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr1_+_233749917
|
10.399
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr20_+_10199455
|
9.943
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr10_+_11060105
|
9.917
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr9_+_130965666
|
9.895
|
|
DNM1
|
dynamin 1
|
|
chr5_+_169064274
|
9.683
|
|
DOCK2
|
dedicator of cytokinesis 2
|
|
chr9_-_139922705
|
9.630
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr2_+_149632772
|
9.459
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr8_+_86376130
|
9.411
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr1_-_85930610
|
9.379
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr11_+_73358593
|
9.155
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr14_-_60097314
|
9.094
|
|
RTN1
|
reticulon 1
|
|
chr16_+_58498182
|
9.045
|
|
NDRG4
|
NDRG family member 4
|
|
chr14_-_60097223
|
8.938
|
|
RTN1
|
reticulon 1
|
|
chr1_+_228675042
|
8.932
|
NM_001010858
|
RNF187
|
ring finger protein 187
|
|
chr1_-_26232890
|
8.700
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr4_+_41258909
|
8.674
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr4_+_41258895
|
8.635
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr2_+_17721806
|
8.510
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr2_+_95691428
|
8.505
|
NM_002371
NM_022438
NM_022439
NM_022440
|
MAL
|
mal, T-cell differentiation protein
|
|
chr4_+_41258941
|
8.488
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr19_-_42498320
|
8.475
|
NM_152296
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr12_+_7023490
|
8.452
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr7_+_75039632
|
8.373
|
|
NSUN5P1
|
NOP2/Sun domain family, member 5 pseudogene 1
|
|
chr2_-_175869910
|
8.290
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr3_+_167453431
|
8.174
|
NM_001122752
NM_005025
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1
|
|
chr15_+_32933869
|
8.160
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr18_-_74844762
|
8.145
|
NM_001025100
NM_001025101
|
MBP
|
myelin basic protein
|
|
chr10_-_15413054
|
8.129
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr16_+_58498216
|
8.076
|
|
NDRG4
|
NDRG family member 4
|
|
chr12_+_122459791
|
8.072
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr19_-_7939325
|
8.061
|
NM_001190467
|
FLJ22184
|
putative uncharacterized protein FLJ22184
|
|
chr3_-_47620186
|
8.058
|
NM_001206943
NM_001206944
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr10_+_105037441
|
7.993
|
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr9_+_130965668
|
7.919
|
|
DNM1
|
dynamin 1
|
|
chr6_-_84419108
|
7.915
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr14_+_104029278
|
7.907
|
NM_032374
|
APOPT1
|
apoptogenic 1
|
|
chr9_+_17578952
|
7.902
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr12_-_46662779
|
7.861
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr16_+_56623266
|
7.849
|
NM_005954
|
MT3
|
metallothionein 3
|
|
chr12_-_46662709
|
7.847
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr17_-_4890664
|
7.833
|
NM_001171167
NM_001171168
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr9_+_130965608
|
7.825
|
NM_001005336
NM_004408
|
DNM1
|
dynamin 1
|
|
chr7_-_37488408
|
7.812
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_+_149571029
|
7.719
|
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr1_+_2005424
|
7.585
|
NM_001242874
|
PRKCZ
|
protein kinase C, zeta
|
|
chr3_+_133494275
|
7.571
|
|
TF
|
transferrin
|
|
chr7_-_37488554
|
7.569
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr20_+_37434294
|
7.533
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr7_-_51384458
|
7.445
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr14_-_103988829
|
7.439
|
|
CKB
|
creatine kinase, brain
|
|
chr16_+_2039945
|
7.433
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr1_-_85930733
|
7.418
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr15_-_38856830
|
7.403
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr2_+_17721983
|
7.361
|
|
VSNL1
|
visinin-like 1
|
|
chr3_+_184056304
|
7.287
|
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr6_+_107811272
|
7.266
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr19_-_42498221
|
7.220
|
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr6_-_46459010
|
7.215
|
NM_001251974
|
RCAN2
|
regulator of calcineurin 2
|
|
chr7_-_30029284
|
7.165
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr12_-_46663207
|
7.151
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr13_-_36705432
|
7.085
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr20_-_3149070
|
7.010
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr14_-_21493910
|
6.923
|
NM_016250
NM_201535
NM_201536
NM_201537
|
NDRG2
|
NDRG family member 2
|
|
chr7_-_44365019
|
6.900
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chrX_-_118827060
|
6.854
|
|
SEPT6
|
septin 6
|
|
chr16_-_2390737
|
6.805
|
NM_001089
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr10_-_118764559
|
6.805
|
|
KIAA1598
|
KIAA1598
|
|
chr14_-_21493858
|
6.804
|
|
NDRG2
|
NDRG family member 2
|
|
chr9_-_91793375
|
6.798
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr14_-_23821423
|
6.798
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr15_+_89182038
|
6.767
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr19_+_35782988
|
6.717
|
NM_001199216
NM_002361
NM_080600
|
MAG
|
myelin associated glycoprotein
|
|
chr2_+_149632836
|
6.677
|
|
KIF5C
|
kinesin family member 5C
|
|
chr22_+_29876180
|
6.594
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr3_+_35681008
|
6.531
|
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr4_-_42658883
|
6.489
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr3_-_18466759
|
6.488
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr16_-_57318574
|
6.448
|
NM_015993
|
PLLP
|
plasmolipin
|
|
chr16_+_14164879
|
6.429
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr17_+_7554239
|
6.410
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr22_-_51021200
|
6.388
|
|
CHKB
|
choline kinase beta
|
|
chr19_+_36359346
|
6.351
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr10_-_98945662
|
6.321
|
NM_003061
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr9_-_136024522
|
6.254
|
NM_001042368
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr2_+_149633173
|
6.226
|
|
|
|
|
chr17_+_7554711
|
6.201
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr9_-_139923214
|
6.178
|
NM_212533
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr9_-_139940595
|
6.176
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
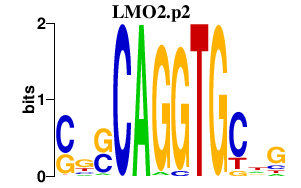
chr11_-_33890931
|
6.153
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr19_+_54926721
|
6.127
|
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr19_-_19006868
|
6.120
|
NM_021267
NM_198207
NM_001492
|
CERS1
GDF1
|
ceramide synthase 1
growth differentiation factor 1
|
|
chr1_+_169075869
|
6.112
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr11_-_695010
|
6.096
|
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr7_-_994259
|
6.083
|
NM_006869
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr3_-_10547340
|
6.055
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr17_+_19912608
|
6.030
|
NM_001243439
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr9_-_139940556
|
6.021
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr9_-_92112887
|
6.020
|
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chrX_+_53111482
|
5.909
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr22_-_50893728
|
5.898
|
|
SBF1
|
SET binding factor 1
|
|
chr6_-_84419018
|
5.837
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr12_-_112819755
|
5.808
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chrX_-_92928566
|
5.808
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr2_-_224702200
|
5.780
|
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr14_-_21493122
|
5.743
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr19_+_5904842
|
5.734
|
NM_001017921
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein
|
|
chr11_+_46354454
|
5.730
|
NM_201532
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr7_-_73123431
|
5.727
|
|
STX1A
|
syntaxin 1A (brain)
|
|
chr8_+_26371461
|
5.688
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr2_-_73496595
|
5.661
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr17_+_16593571
|
5.653
|
NM_014695
|
CCDC144B
CCDC144A
|
coiled-coil domain containing 144B (pseudogene)
coiled-coil domain containing 144A
|
|
chr22_+_38453478
|
5.618
|
|
PICK1
|
protein interacting with PRKCA 1
|
|
chr5_-_179285791
|
5.613
|
NM_001017987
NM_016175
|
C5orf45
|
chromosome 5 open reading frame 45
|
|
chr14_+_96505566
|
5.606
|
NM_001252507
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr3_-_49907304
|
5.588
|
NM_024046
|
CAMKV
|
CaM kinase-like vesicle-associated
|
|
chr19_-_58609606
|
5.502
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr5_+_173472737
|
5.480
|
|
HMP19
|
HMP19 protein
|
|
chr12_+_7342956
|
5.478
|
NM_000319
|
PEX5
|
peroxisomal biogenesis factor 5
|
|
chr7_-_72425211
|
5.476
|
|
NSUN5P1
NSUN5P2
|
NOP2/Sun domain family, member 5 pseudogene 1
NOP2/Sun domain family, member 5 pseudogene 2
|
|
chr9_-_139940464
|
5.470
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr3_-_120068077
|
5.351
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr13_+_113656027
|
5.310
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr7_-_72722786
|
5.296
|
NM_001168347
NM_001168348
NM_018044
NM_148956
|
NSUN5
|
NOP2/Sun domain family, member 5
|
|
chr14_+_105781180
|
5.290
|
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr1_+_6094342
|
5.285
|
NM_001199860
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr7_-_143059144
|
5.279
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr16_+_7382750
|
5.262
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr7_-_124405680
|
5.239
|
NM_005302
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr17_-_4890920
|
5.213
|
NM_001171166
NM_015099
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr9_+_87284625
|
5.203
|
NM_001007097
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chrX_-_118827322
|
5.188
|
NM_015129
NM_145799
NM_145800
NM_145802
|
SEPT6
|
septin 6
|
|
chr16_-_2390711
|
5.180
|
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr1_-_233431458
|
5.164
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr19_-_36523540
|
5.106
|
NM_001199570
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr1_-_226595533
|
5.063
|
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr2_+_173600634
|
5.045
|
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr9_-_10612722
|
5.044
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr22_+_32870829
|
5.024
|
|
FBXO7
|
F-box protein 7
|
|
chr10_-_46342906
|
5.012
|
NM_133446
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4
|
|
chr1_+_78354199
|
5.011
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr8_-_103137134
|
5.009
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr8_-_103136486
|
4.982
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr1_-_40782981
|
4.969
|
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr17_+_43299277
|
4.958
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr5_-_88199868
|
4.944
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr17_-_79139871
|
4.938
|
NM_001080395
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr11_-_72380107
|
4.935
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr10_+_11060069
|
4.910
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_41131323
|
4.909
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|