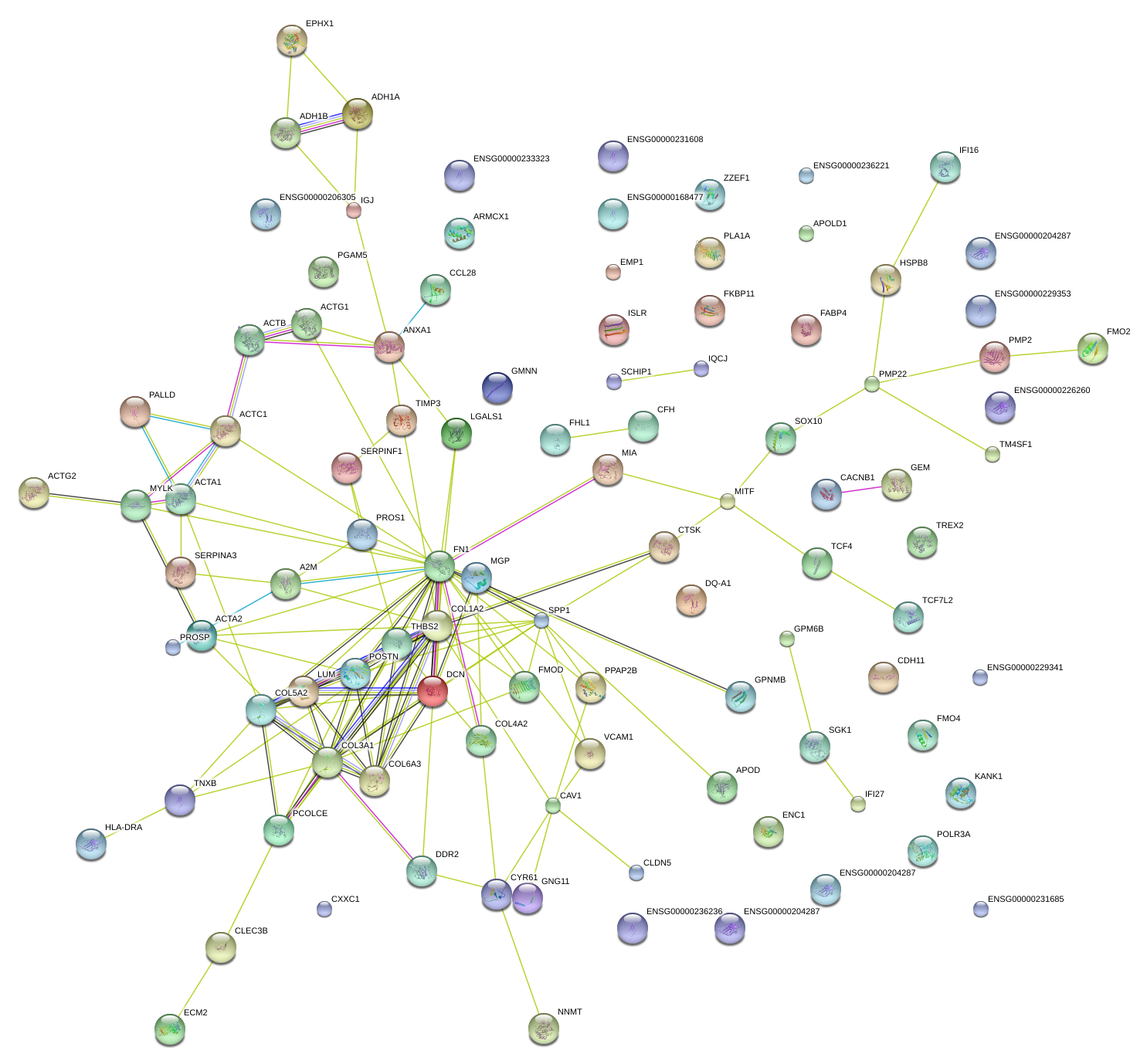
|
chr2_-_238322840
|
46.223
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_238322770
|
44.200
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_+_189839132
|
43.950
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_-_238322815
|
36.100
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_+_189839078
|
34.846
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189839113
|
33.063
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_-_91573264
|
27.767
|
NM_133503
|
DCN
|
decorin
|
|
chr12_+_13349720
|
21.734
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_+_13349601
|
21.414
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr2_-_216259197
|
19.664
|
|
FN1
|
fibronectin 1
|
|
chr7_+_23286384
|
17.397
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286386
|
17.255
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286390
|
17.240
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr12_-_15038724
|
15.951
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr17_-_15164043
|
15.921
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr1_-_150780705
|
14.013
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr3_-_195310985
|
13.867
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr6_-_169653922
|
13.595
|
|
THBS2
|
thrombospondin 2
|
|
chr12_-_9268513
|
13.379
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr7_+_116164838
|
13.341
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr1_+_162602308
|
13.024
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr1_+_162602198
|
12.963
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr6_-_169654047
|
12.904
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr22_+_33196801
|
12.118
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr13_+_111134933
|
11.565
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr7_+_23286277
|
11.347
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_+_86046434
|
11.245
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr5_-_42811959
|
11.240
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr1_-_203320249
|
11.134
|
|
|
|
|
chr6_-_134639195
|
11.068
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr7_+_94024201
|
10.801
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr3_-_195310770
|
10.557
|
|
APOD
|
apolipoprotein D
|
|
chr6_-_134495944
|
10.453
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr18_-_53068755
|
10.298
|
|
TCF4
|
transcription factor 4
|
|
chr3_+_119316694
|
10.296
|
NM_001206960
NM_001206961
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr12_-_9268475
|
10.119
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr3_-_123411185
|
10.050
|
|
MYLK
|
myosin light chain kinase
|
|
chr1_-_203320287
|
9.987
|
NM_002023
|
FMOD
|
fibromodulin
|
|
chr12_-_9268522
|
9.914
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr6_+_32407618
|
9.838
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr1_-_203320185
|
9.783
|
|
FMOD
|
fibromodulin
|
|
chr7_+_23286420
|
9.776
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_+_226016428
|
9.717
|
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr2_-_216241321
|
9.303
|
|
FN1
|
fibronectin 1
|
|
chr4_+_169418167
|
9.119
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr1_+_101185195
|
9.020
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr6_-_134496006
|
8.880
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr7_+_93551015
|
8.684
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr12_-_9268440
|
8.651
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr12_-_91505215
|
8.130
|
|
LUM
|
lumican
|
|
chr12_+_12938540
|
8.102
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr3_+_159557745
|
7.728
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr4_-_71532206
|
7.706
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr9_-_95298251
|
7.594
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr1_+_158979681
|
7.478
|
NM_001206567
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr14_+_94577078
|
7.355
|
NM_001130080
NM_005532
|
IFI27
|
interferon, alpha-inducible protein 27
|
|
chr7_-_64023444
|
7.323
|
|
|
|
|
chr4_-_71532341
|
7.262
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr22_+_38071612
|
7.259
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr2_-_89165652
|
7.224
|
|
|
|
|
chr8_-_82395401
|
7.201
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr5_-_43412417
|
7.028
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr14_+_95078713
|
6.973
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr12_-_91505541
|
6.779
|
NM_002345
|
LUM
|
lumican
|
|
chr3_-_93692898
|
6.757
|
NM_000313
|
PROS1
|
protein S (alpha)
|
|
chr3_+_69812961
|
6.709
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_+_88896868
|
6.660
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr9_+_706744
|
6.439
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr4_+_88896899
|
6.433
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chrX_-_13835313
|
6.362
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr4_-_100242487
|
6.362
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr9_+_75766777
|
6.352
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr8_-_95274520
|
6.248
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr8_-_95274540
|
6.166
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr12_+_119616594
|
6.157
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr3_-_149095413
|
6.155
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr8_-_82359620
|
6.118
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr4_+_88896838
|
5.924
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr6_-_31981049
|
5.873
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr3_-_195306274
|
5.853
|
|
APOD
|
apolipoprotein D
|
|
chr13_-_38172862
|
5.834
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr3_+_45067750
|
5.755
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr3_-_149095517
|
5.733
|
NM_014220
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr1_-_57045235
|
5.730
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr16_-_65155881
|
5.726
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr10_-_90712510
|
5.644
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr3_-_149095333
|
5.615
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr2_-_190044330
|
5.613
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chrX_-_13835187
|
5.567
|
|
GPM6B
|
glycoprotein M6B
|
|
chr22_-_38380538
|
5.567
|
NM_006941
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chr22_-_19512859
|
5.544
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr5_+_140739589
|
5.540
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr7_+_100199881
|
5.472
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr11_+_114167197
|
5.466
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chrX_+_135229536
|
5.459
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr1_+_171154387
|
5.438
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chrX_-_13835012
|
5.381
|
|
GPM6B
|
glycoprotein M6B
|
|
chrX_+_135251454
|
5.338
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr3_-_149095458
|
5.327
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr15_+_74466024
|
5.228
|
NM_005545
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr17_+_1665281
|
5.158
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr19_+_41281081
|
5.045
|
NM_001202553
|
MIA
|
melanoma inhibitory activity
|
|
chr12_-_91576805
|
4.992
|
NM_001920
|
DCN
|
decorin
|
|
chr10_-_79789290
|
4.913
|
NM_007055
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa
|
|
chrX_+_100805415
|
4.787
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr12_-_91576579
|
4.753
|
|
DCN
|
decorin
|
|
chr4_+_71248794
|
4.744
|
NM_006685
|
SMR3B
|
submaxillary gland androgen regulated protein 3B
|
|
chr7_-_100860991
|
4.649
|
NM_001084
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr12_-_8693391
|
4.643
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr14_-_107131217
|
4.641
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr6_-_131384387
|
4.615
|
NM_001135554
NM_001135555
NM_001199388
NM_001431
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr14_-_106573348
|
4.614
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chrX_+_103031753
|
4.547
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr5_+_34684611
|
4.546
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr18_-_52988916
|
4.539
|
NM_001243234
NM_001243235
|
TCF4
|
transcription factor 4
|
|
chr5_+_140729723
|
4.511
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr4_-_76928601
|
4.511
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr5_+_140868721
|
4.490
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr19_+_16222706
|
4.430
|
|
RAB8A
|
RAB8A, member RAS oncogene family
|
|
chr1_+_159175048
|
4.428
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr12_-_50677238
|
4.425
|
NM_001113546
NM_016357
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr3_+_361365
|
4.424
|
NM_001253388
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr19_+_41281281
|
4.400
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr20_+_19915716
|
4.369
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr14_-_92413738
|
4.353
|
|
FBLN5
|
fibulin 5
|
|
chr9_-_35689799
|
4.344
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr11_+_57365026
|
4.343
|
NM_000062
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr11_-_8892909
|
4.343
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr8_-_49833823
|
4.313
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr20_+_19867075
|
4.293
|
NM_001242581
|
RIN2
|
Ras and Rab interactor 2
|
|
chr10_+_17851342
|
4.286
|
NM_002438
|
MRC1
|
mannose receptor, C type 1
|
|
chrX_+_103031797
|
4.285
|
|
PLP1
|
proteolipid protein 1
|
|
chr4_+_88896801
|
4.244
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr16_+_1584161
|
4.224
|
NM_024600
|
TMEM204
|
transmembrane protein 204
|
|
chr4_+_74269971
|
4.206
|
NM_000477
|
ALB
|
albumin
|
|
chr9_-_13279562
|
4.192
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chr4_+_175204827
|
4.188
|
NM_001040157
|
CEP44
|
centrosomal protein 44kDa
|
|
chr21_-_28339405
|
4.184
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr4_+_175205044
|
4.172
|
NM_001145314
|
CEP44
|
centrosomal protein 44kDa
|
|
chr3_-_137893718
|
4.151
|
|
DBR1
|
debranching enzyme homolog 1 (S. cerevisiae)
|
|
chr11_-_65382701
|
4.125
|
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr9_-_35689902
|
4.121
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr19_+_37998041
|
4.119
|
|
ZNF793
|
zinc finger protein 793
|
|
chr1_-_169599349
|
4.119
|
NM_003005
|
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62)
|
|
chr11_+_308106
|
4.118
|
NM_006435
|
IFITM2
|
interferon induced transmembrane protein 2 (1-8D)
|
|
chr3_+_186560462
|
4.117
|
NM_001177800
NM_004797
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr17_+_32582295
|
4.078
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr6_-_112575880
|
4.062
|
|
LAMA4
|
laminin, alpha 4
|
|
chr1_-_163113938
|
4.045
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr14_-_106926393
|
4.041
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_+_27301693
|
4.028
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr5_-_77844975
|
3.980
|
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr3_-_93692592
|
3.964
|
|
PROS1
|
protein S (alpha)
|
|
chr9_+_75766794
|
3.963
|
|
ANXA1
|
annexin A1
|
|
chr10_-_7682741
|
3.947
|
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr3_+_186560478
|
3.945
|
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr15_-_45480160
|
3.934
|
|
SHF
|
Src homology 2 domain containing F
|
|
chr3_-_183273407
|
3.931
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr2_+_99797632
|
3.925
|
|
MRPL30
|
mitochondrial ribosomal protein L30
|
|
chr1_-_94702916
|
3.911
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr1_+_203595929
|
3.853
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr12_+_54422193
|
3.848
|
NM_004503
|
HOXC6
|
homeobox C6
|
|
chr14_-_106746056
|
3.842
|
|
|
|
|
chr1_-_32169555
|
3.817
|
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr11_-_10590078
|
3.801
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chrX_+_152760346
|
3.795
|
NM_001711
|
BGN
|
biglycan
|
|
chr5_-_146833150
|
3.794
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chrX_+_152760452
|
3.782
|
|
BGN
|
biglycan
|
|
chr6_+_116692098
|
3.759
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr1_-_32169619
|
3.745
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr19_+_41509825
|
3.699
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr22_-_38379868
|
3.676
|
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chr17_-_40575117
|
3.673
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr4_+_124317884
|
3.662
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr6_-_112575766
|
3.657
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr17_-_48262900
|
3.646
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr8_+_17433941
|
3.588
|
NM_006207
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chr14_-_69259654
|
3.583
|
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr12_+_69742140
|
3.557
|
|
LYZ
|
lysozyme
|
|
chr9_-_95166827
|
3.536
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr12_-_7245048
|
3.535
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr1_+_99127146
|
3.497
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chr17_-_56160564
|
3.494
|
|
|
|
|
chr6_-_46825942
|
3.488
|
|
GPR116
|
G protein-coupled receptor 116
|
|
chr14_+_55595966
|
3.468
|
|
LGALS3
|
lectin, galactoside-binding, soluble, 3
|
|
chr13_-_45150391
|
3.443
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr2_+_99797536
|
3.431
|
NM_145212
|
MRPL30
|
mitochondrial ribosomal protein L30
|
|
chr9_-_129884998
|
3.426
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr1_-_68697997
|
3.426
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr16_+_84853585
|
3.416
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr6_-_131384321
|
3.412
|
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr16_-_3306586
|
3.405
|
NM_000243
NM_001198536
|
MEFV
|
Mediterranean fever
|
|
chr6_+_101846668
|
3.366
|
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr2_-_97405799
|
3.354
|
NM_001142292
NM_030805
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr6_-_32013806
|
3.353
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr5_+_140810181
|
3.344
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr6_-_46922533
|
3.344
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr1_-_94703252
|
3.340
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr2_-_97405775
|
3.333
|
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr2_-_56151273
|
3.320
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|