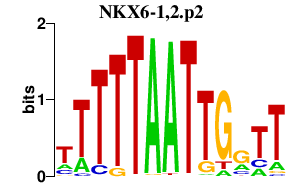
|
chr4_+_169418167
|
15.770
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr3_-_79817058
|
12.583
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr8_+_132991171
|
12.167
|
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr7_+_94297448
|
8.988
|
|
PEG10
|
paternally expressed 10
|
|
chr7_+_23286420
|
8.578
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr2_-_238322840
|
8.458
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr7_+_23286384
|
8.440
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_+_84647342
|
7.991
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr6_+_87969619
|
7.716
|
|
ZNF292
|
zinc finger protein 292
|
|
chr7_-_137686814
|
7.434
|
NM_001253775
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr7_+_23286386
|
7.388
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr2_-_238322815
|
7.316
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr5_+_140729723
|
6.915
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr7_+_23286390
|
6.705
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chrX_+_80457302
|
6.285
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr12_-_91505541
|
6.133
|
NM_002345
|
LUM
|
lumican
|
|
chr20_+_11898536
|
6.084
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chrX_+_102840412
|
5.848
|
NM_001006935
NM_001006937
NM_024863
|
TCEAL4
|
transcription elongation factor A (SII)-like 4
|
|
chr1_-_226926875
|
5.838
|
NM_002221
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr12_-_91505215
|
5.766
|
|
LUM
|
lumican
|
|
chr7_-_137028533
|
5.737
|
NM_002825
|
PTN
|
pleiotrophin
|
|
chr4_+_70894129
|
5.635
|
NM_000200
|
HTN3
|
histatin 3
|
|
chr20_+_19915716
|
5.607
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_+_104159953
|
5.550
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr4_+_88896801
|
5.548
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr7_+_23286277
|
5.531
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_-_93342706
|
5.498
|
NM_001252271
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chrX_-_10535476
|
5.494
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chrX_+_47441650
|
5.148
|
NM_003254
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr2_-_151344145
|
4.945
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr4_-_186732047
|
4.821
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_+_140739589
|
4.796
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr4_-_186733372
|
4.769
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_114178516
|
4.625
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr6_-_75970566
|
4.619
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr4_-_83347222
|
4.603
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr18_-_51690988
|
4.597
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr2_-_175712276
|
4.497
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr4_+_166299942
|
4.462
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr4_+_88896899
|
4.454
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chrX_+_47441802
|
4.288
|
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chrX_-_100872926
|
4.178
|
NM_001009584
NM_001184768
NM_019007
|
ARMCX6
|
armadillo repeat containing, X-linked 6
|
|
chr15_+_25068793
|
4.171
|
NM_022806
NM_022807
NM_022808
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr11_+_17332454
|
4.101
|
|
|
|
|
chr2_+_108994366
|
4.092
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr4_+_55095263
|
4.023
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr10_+_63808968
|
3.965
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr1_-_21377452
|
3.920
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr7_-_26232148
|
3.913
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr3_-_114790221
|
3.781
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr1_-_115269672
|
3.697
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr1_+_104097321
|
3.694
|
NM_020978
|
AMY2B
|
amylase, alpha 2B (pancreatic)
|
|
chr4_+_41700736
|
3.614
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr22_-_26961333
|
3.587
|
NM_001008566
|
TPST2
|
tyrosylprotein sulfotransferase 2
|
|
chr8_+_26150731
|
3.543
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr4_+_83956238
|
3.453
|
NM_016129
|
COPS4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis)
|
|
chr4_+_71337674
|
3.428
|
NM_001145007
|
MUC7
|
mucin 7, secreted
|
|
chr4_+_41258895
|
3.416
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr1_+_53480616
|
3.367
|
|
SCP2
|
sterol carrier protein 2
|
|
chr2_-_47134945
|
3.359
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr12_+_119616594
|
3.322
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr3_-_15382874
|
3.307
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr3_-_105421044
|
3.275
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr3_-_149421059
|
3.247
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chrX_-_114252132
|
3.245
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr6_+_158733691
|
3.227
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr12_-_6233684
|
3.195
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr13_-_38172862
|
3.154
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr12_-_46633551
|
3.152
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr1_-_207095198
|
3.117
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr12_-_54653317
|
3.096
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chrX_-_10544852
|
3.083
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr10_-_98321823
|
3.063
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr6_+_26440699
|
3.060
|
NM_001242803
NM_006994
NM_197974
|
BTN3A3
|
butyrophilin, subfamily 3, member A3
|
|
chrX_+_80457597
|
3.051
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr4_+_71337833
|
3.048
|
NM_152291
|
MUC7
|
mucin 7, secreted
|
|
chrX_-_23855456
|
2.993
|
|
RPL9
|
ribosomal protein L9
|
|
chr1_+_53480573
|
2.942
|
NM_001007099
NM_001007100
NM_001007250
|
SCP2
|
sterol carrier protein 2
|
|
chr5_+_67586466
|
2.914
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr12_-_54653369
|
2.910
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr1_-_21377387
|
2.874
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr1_+_84630374
|
2.846
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr7_-_22862397
|
2.826
|
NM_019059
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast)
|
|
chr9_+_706744
|
2.772
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr5_-_150727088
|
2.741
|
NM_181776
|
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2
|
|
chr12_+_9980076
|
2.730
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr5_-_111091947
|
2.696
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr4_+_41258909
|
2.690
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chrX_+_70765479
|
2.640
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr6_+_26365397
|
2.562
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr3_+_23851933
|
2.560
|
NM_001202476
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1
|
|
chr18_-_47018777
|
2.543
|
|
RPL17
|
ribosomal protein L17
|
|
chr2_-_10942642
|
2.524
|
|
PDIA6
|
protein disulfide isomerase family A, member 6
|
|
chr4_-_71532206
|
2.478
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr2_+_183608329
|
2.448
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr20_-_20036685
|
2.442
|
NM_016652
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr12_+_28410132
|
2.438
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr4_-_16900028
|
2.425
|
|
LDB2
|
LIM domain binding 2
|
|
chr4_+_88896838
|
2.423
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr12_+_62954275
|
2.416
|
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr4_+_88896868
|
2.415
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr5_+_140749830
|
2.398
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr13_-_33760215
|
2.388
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_+_68100994
|
2.386
|
NM_170742
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr2_+_109204904
|
2.385
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_+_109384601
|
2.384
|
|
RANBP2
|
RAN binding protein 2
|
|
chr9_+_82186846
|
2.375
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr1_+_104198301
|
2.346
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr14_-_95599794
|
2.305
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr3_+_63884074
|
2.288
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr4_+_71248794
|
2.278
|
NM_006685
|
SMR3B
|
submaxillary gland androgen regulated protein 3B
|
|
chr18_-_47018842
|
2.243
|
NM_001199356
NM_000985
NM_001035006
NM_001199340
NM_001199345
|
RPL17-C18ORF32
RPL17
|
RPL17-C18orf32 readthrough
ribosomal protein L17
|
|
chrX_-_78623006
|
2.236
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chrX_-_10851808
|
2.208
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chrX_-_33229428
|
2.167
|
NM_004006
|
DMD
|
dystrophin
|
|
chr1_-_6420719
|
2.166
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr4_-_186877413
|
2.152
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr8_+_79428335
|
2.127
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr7_-_137028275
|
2.126
|
|
PTN
|
pleiotrophin
|
|
chr2_+_223725675
|
2.118
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr1_+_104292439
|
2.109
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chrX_+_135230736
|
2.105
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr10_+_28878666
|
2.102
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr2_-_217560247
|
2.053
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr9_+_116263706
|
2.049
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr22_-_39268214
|
1.984
|
|
CBX6
|
chromobox homolog 6
|
|
chr9_+_34652125
|
1.966
|
NM_001142784
|
IL11RA
|
interleukin 11 receptor, alpha
|
|
chr22_-_39268169
|
1.963
|
|
CBX6
|
chromobox homolog 6
|
|
chr18_-_47018822
|
1.962
|
NM_001199341
NM_001199342
NM_001199343
NM_001199344
|
RPL17
|
ribosomal protein L17
|
|
chr3_-_149095413
|
1.961
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr1_-_110933635
|
1.933
|
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr1_+_100316530
|
1.919
|
NM_000646
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
|
|
chr17_-_17480325
|
1.906
|
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr4_-_16900256
|
1.897
|
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_16900348
|
1.875
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr15_+_44092618
|
1.870
|
NM_001199885
NM_016400
|
C15orf63
|
chromosome 15 open reading frame 63
|
|
chr2_+_172821874
|
1.861
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr11_+_7618416
|
1.830
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr6_+_148663728
|
1.829
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr5_+_140868721
|
1.828
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr1_+_79115476
|
1.786
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr7_-_41742666
|
1.778
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr19_+_41281281
|
1.777
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr1_+_84630575
|
1.767
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr2_-_37544165
|
1.738
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr22_-_39268230
|
1.734
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr1_-_197552304
|
1.720
|
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chr5_+_95066628
|
1.714
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr2_-_74382352
|
1.680
|
|
|
|
|
chr13_-_45048436
|
1.668
|
NM_001243798
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr13_+_52598678
|
1.642
|
NM_021645
|
ALG11
UTP14C
|
asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast)
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast)
|
|
chr12_+_7023490
|
1.635
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr4_+_169013687
|
1.591
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr6_+_43738394
|
1.555
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr5_+_140810181
|
1.533
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr12_+_10457044
|
1.525
|
NM_001114396
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr6_-_87804773
|
1.524
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr4_-_110723334
|
1.519
|
NM_000204
|
CFI
|
complement factor I
|
|
chr5_+_42548144
|
1.514
|
NM_001242402
|
GHR
|
growth hormone receptor
|
|
chr1_+_84609941
|
1.493
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr5_+_49961732
|
1.449
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr17_-_7307413
|
1.445
|
NM_152766
|
C17orf61-PLSCR3
C17orf61
|
C17orf61-PLSCR3 readthrough
chromosome 17 open reading frame 61
|
|
chr2_-_169103886
|
1.445
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr13_-_110802708
|
1.444
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr11_-_115088628
|
1.435
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr13_+_53602829
|
1.426
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr2_+_228356287
|
1.419
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr12_-_53600999
|
1.415
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr22_+_31366662
|
1.394
|
|
|
|
|
chr22_-_43411129
|
1.392
|
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr3_+_132036210
|
1.389
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr8_+_17822093
|
1.388
|
|
PCM1
|
pericentriolar material 1
|
|
chr5_+_42565562
|
1.385
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr6_+_26402464
|
1.370
|
NM_001145008
NM_001145009
NM_007048
NM_194441
|
BTN3A1
|
butyrophilin, subfamily 3, member A1
|
|
chr1_+_79086087
|
1.365
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr22_-_38881529
|
1.345
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr10_+_111765725
|
1.340
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr3_-_149095333
|
1.339
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr17_+_68071386
|
1.333
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr5_+_140810126
|
1.332
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr3_-_119582422
|
1.330
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr11_-_67205151
|
1.321
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chrX_+_123480131
|
1.316
|
NM_001114937
NM_002351
|
SH2D1A
|
SH2 domain containing 1A
|
|
chr7_-_137028370
|
1.305
|
|
PTN
|
pleiotrophin
|
|
chr4_+_41258941
|
1.301
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr12_-_53601051
|
1.287
|
|
ITGB7
|
integrin, beta 7
|
|
chr17_+_8191814
|
1.282
|
NM_001177801
NM_001177802
NM_016492
|
RANGRF
|
RAN guanine nucleotide release factor
|
|
chr5_+_65372744
|
1.274
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr9_+_97488943
|
1.264
|
NM_001193331
NM_032823
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr18_+_72166822
|
1.262
|
NM_001168499
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr3_-_58563070
|
1.261
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr4_-_186877869
|
1.249
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_+_109204745
|
1.242
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr10_-_98325100
|
1.238
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr10_+_86131546
|
1.227
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr1_+_146649544
|
1.222
|
|
|
|
|
chr15_-_66648991
|
1.220
|
NM_017858
|
TIPIN
|
TIMELESS interacting protein
|
|
chr12_+_118573945
|
1.206
|
|
PEBP1
|
phosphatidylethanolamine binding protein 1
|
|
chr9_+_97488993
|
1.205
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr5_+_179132676
|
1.194
|
|
CANX
|
calnexin
|