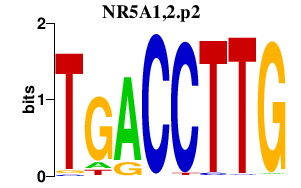
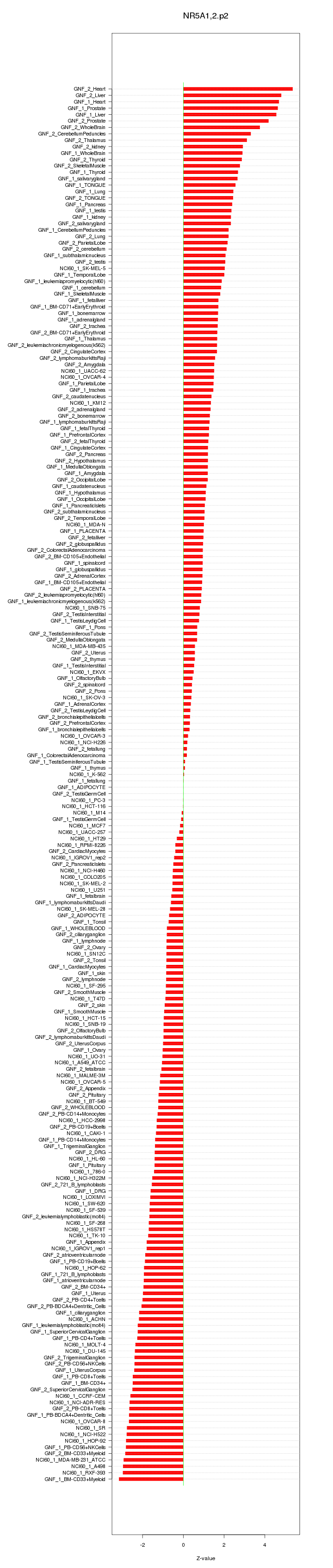
|
chr1_+_55464605
|
32.193
|
NM_057176
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin)
|
|
chr11_-_5275904
|
27.279
|
NM_000184
|
HBG1
HBG2
|
hemoglobin, gamma A
hemoglobin, gamma G
|
|
chr11_-_5271035
|
27.168
|
NM_000559
|
HBG1
|
hemoglobin, gamma A
|
|
chr10_-_101190201
|
26.008
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr22_+_41865112
|
23.866
|
NM_001098
|
ACO2
|
aconitase 2, mitochondrial
|
|
chrX_+_103031433
|
23.819
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chrX_+_103031797
|
23.555
|
|
PLP1
|
proteolipid protein 1
|
|
chrX_+_103031753
|
21.437
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr12_-_120554558
|
20.871
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr12_-_9268440
|
19.637
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr12_-_9268522
|
19.417
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr6_-_108145507
|
18.706
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr12_-_9268513
|
18.669
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr2_-_218843541
|
18.363
|
|
TNS1
|
tensin 1
|
|
chr14_+_93389491
|
17.745
|
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr5_-_42811959
|
17.507
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr17_-_48271968
|
16.967
|
|
|
|
|
chr5_-_11904127
|
16.271
|
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr2_+_98262497
|
15.395
|
NM_001862
|
COX5B
|
cytochrome c oxidase subunit Vb
|
|
chr3_+_133464883
|
14.635
|
NM_001063
|
TF
|
transferrin
|
|
chr16_+_226678
|
14.491
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr12_-_9268475
|
14.336
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr14_-_103989195
|
14.295
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr14_-_103989112
|
14.133
|
|
CKB
|
creatine kinase, brain
|
|
chr1_-_182354649
|
13.690
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr3_+_133465250
|
13.630
|
|
TF
|
transferrin
|
|
chr3_+_133465098
|
13.594
|
|
TF
|
transferrin
|
|
chr5_+_80529138
|
13.546
|
NM_001099735
NM_001099736
NM_001825
|
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric)
|
|
chr5_-_11903964
|
13.479
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr12_-_106532447
|
13.445
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chrX_-_13835187
|
13.225
|
|
GPM6B
|
glycoprotein M6B
|
|
chr11_-_5248180
|
13.124
|
NM_000518
|
HBB
|
hemoglobin, beta
|
|
chr3_+_133465204
|
13.072
|
|
TF
|
transferrin
|
|
chr12_+_119616594
|
12.214
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr19_-_36643770
|
12.156
|
NM_001864
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle)
|
|
chr14_-_103988829
|
12.017
|
|
CKB
|
creatine kinase, brain
|
|
chr14_-_106174914
|
11.738
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr2_-_175869910
|
11.652
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr15_+_91769329
|
11.500
|
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chrX_-_13835012
|
11.475
|
|
GPM6B
|
glycoprotein M6B
|
|
chr18_-_74728963
|
11.410
|
|
MBP
|
myelin basic protein
|
|
chr12_+_48516428
|
11.302
|
NM_001166687
NM_001166688
|
PFKM
|
phosphofructokinase, muscle
|
|
chr19_-_57336373
|
11.277
|
NM_001146186
|
PEG3
|
paternally expressed 3
|
|
chr18_-_74729049
|
11.172
|
NM_001025081
NM_001025090
NM_001025092
NM_002385
|
MBP
|
myelin basic protein
|
|
chr17_-_48271365
|
11.044
|
|
|
|
|
chr17_-_26903854
|
10.653
|
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr22_-_19511777
|
10.541
|
|
CLDN5
|
claudin 5
|
|
chr12_-_11463353
|
10.465
|
NM_002723
|
PRB4
|
proline-rich protein BstNI subfamily 4
|
|
chr12_-_120554551
|
10.218
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr21_-_27107184
|
10.072
|
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6
|
|
chr22_-_44258210
|
10.028
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr19_-_45826131
|
9.826
|
|
CKM
|
creatine kinase, muscle
|
|
chr8_-_120651019
|
9.535
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr3_-_47957475
|
9.517
|
|
|
|
|
chr8_-_22089517
|
9.462
|
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr20_-_32031425
|
9.410
|
NM_003098
|
SNTA1
|
syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component)
|
|
chr12_-_120554453
|
9.398
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr6_+_30856462
|
9.361
|
NM_001202521
NM_001202522
NM_013994
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr8_+_136469557
|
9.333
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr14_+_93389444
|
9.325
|
NM_001275
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr12_-_122296619
|
9.306
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr6_-_46293628
|
9.267
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr11_+_73359734
|
9.057
|
NM_001130036
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr12_-_11002051
|
9.050
|
NM_001098538
NM_007244
|
PRR4
|
proline rich 4 (lacrimal)
|
|
chrX_-_107225588
|
8.923
|
NM_031273
|
TEX13B
|
testis expressed 13B
|
|
chr22_+_23040394
|
8.906
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr7_-_44105162
|
8.689
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr16_+_56623266
|
8.579
|
NM_005954
|
MT3
|
metallothionein 3
|
|
chr16_+_2587989
|
8.571
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr3_-_52312257
|
8.492
|
|
WDR82
|
WD repeat domain 82
|
|
chr16_+_2588000
|
8.481
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr12_-_111358353
|
8.439
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr1_+_2036154
|
8.372
|
NM_001033582
|
PRKCZ
|
protein kinase C, zeta
|
|
chr11_-_790103
|
8.254
|
NM_016564
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr3_-_123339423
|
8.249
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr17_-_26903926
|
8.225
|
NM_005165
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr1_+_236849753
|
8.217
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr10_+_88428167
|
8.079
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr12_+_109577201
|
7.938
|
NM_001093
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr20_-_62130390
|
7.908
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr3_-_50605062
|
7.901
|
NM_001171740
NM_001171743
NM_016210
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr11_-_66639602
|
7.898
|
|
|
|
|
chr16_+_2588021
|
7.820
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr22_+_23222897
|
7.807
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr11_-_62472901
|
7.798
|
|
|
|
|
chr17_-_8066254
|
7.789
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chrX_+_70443055
|
7.779
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr8_-_110657025
|
7.755
|
NM_001099753
NM_001099754
NM_001099755
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr17_+_26800657
|
7.724
|
NM_001145975
NM_001145976
NM_003984
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2
|
|
chr3_-_123339178
|
7.716
|
|
MYLK
|
myosin light chain kinase
|
|
chr12_-_11422640
|
7.612
|
NM_006249
|
PRB3
|
proline-rich protein BstNI subfamily 3
|
|
chr22_-_36019363
|
7.595
|
NM_203377
|
MB
|
myoglobin
|
|
chr6_+_27775898
|
7.591
|
NM_003509
|
HIST1H2AI
HIST1H2AM
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H2am
histone cluster 1, H3f
|
|
chr16_+_57481336
|
7.588
|
NM_020312
|
COQ9
|
coenzyme Q9 homolog (S. cerevisiae)
|
|
chr5_+_161274196
|
7.569
|
NM_000806
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr17_-_56618178
|
7.538
|
NM_001198713
|
SEPT4
|
septin 4
|
|
chr3_+_50306578
|
7.520
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr17_+_7608413
|
7.516
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr4_-_57522469
|
7.338
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr2_+_220492286
|
7.314
|
NM_005070
NM_201574
|
SLC4A3
|
solute carrier family 4, anion exchanger, member 3
|
|
chr22_-_39240016
|
7.272
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr14_-_106237486
|
7.194
|
|
|
|
|
chr22_-_29711650
|
7.192
|
NM_001007279
NM_006477
|
RASL10A
|
RAS-like, family 10, member A
|
|
chr14_-_107013474
|
7.185
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr22_-_29711474
|
7.176
|
|
RASL10A
|
RAS-like, family 10, member A
|
|
chr5_+_140868721
|
7.152
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr17_-_73505914
|
7.141
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr3_+_52813936
|
7.128
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr3_+_49027281
|
7.125
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr8_+_86376130
|
7.122
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr7_-_124405680
|
7.072
|
NM_005302
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr1_+_1260289
|
7.059
|
|
|
|
|
chr12_-_16759531
|
7.045
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr19_+_19626535
|
6.968
|
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13
|
|
chr2_-_242556883
|
6.943
|
NM_001164356
|
THAP4
|
THAP domain containing 4
|
|
chr4_-_88450243
|
6.939
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr4_+_30722191
|
6.935
|
|
PCDH7
|
protocadherin 7
|
|
chr16_-_4852571
|
6.906
|
NM_024589
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr19_+_45449228
|
6.887
|
NM_000483
|
APOC2
|
apolipoprotein C-II
|
|
chr5_-_134914968
|
6.845
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr1_-_171621750
|
6.837
|
NM_000261
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response
|
|
chr9_+_126773851
|
6.826
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr10_+_105036783
|
6.823
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr20_-_23669606
|
6.818
|
NM_001899
|
CST4
|
cystatin S
|
|
chr6_-_27782517
|
6.775
|
NM_021066
|
HIST1H2AJ
|
histone cluster 1, H2aj
|
|
chr6_+_27100784
|
6.765
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr1_+_19923457
|
6.723
|
NM_001204088
NM_001204089
NM_001032363
NM_001204082
NM_001204083
|
C1orf151-NBL1
MINOS1
|
C1orf151-NBL1 readthrough
mitochondrial inner membrane organizing system 1
|
|
chr2_-_179672056
|
6.708
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr20_-_48532036
|
6.686
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr22_+_30792929
|
6.657
|
NM_001204204
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr9_+_139871954
|
6.657
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr8_-_42397048
|
6.650
|
NM_006749
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chr1_+_104198140
|
6.636
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr20_-_48531986
|
6.633
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr17_-_39743085
|
6.616
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr12_-_10151689
|
6.602
|
NM_001099431
NM_016509
|
CLEC1B
|
C-type lectin domain family 1, member B
|
|
chr22_-_38379868
|
6.584
|
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chr7_+_153584383
|
6.563
|
NM_001039350
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr2_-_45236521
|
6.551
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr1_+_165600433
|
6.544
|
|
MGST3
|
microsomal glutathione S-transferase 3
|
|
chr11_-_5255712
|
6.515
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr1_-_12677347
|
6.509
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr1_+_104198301
|
6.492
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr16_-_1877140
|
6.478
|
NM_001040427
NM_005326
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr3_+_52812438
|
6.445
|
NM_001166434
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr7_+_29519485
|
6.417
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr19_-_55668956
|
6.388
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr3_-_112280759
|
6.369
|
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr22_-_36013303
|
6.360
|
NM_005368
NM_203378
|
MB
|
myoglobin
|
|
chr9_+_17578952
|
6.349
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr14_+_100150596
|
6.326
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr1_-_57045214
|
6.291
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr6_-_27806116
|
6.289
|
NM_003510
|
HIST1H2AK
|
histone cluster 1, H2ak
|
|
chr17_+_7211260
|
6.270
|
NM_001143761
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr1_-_182922002
|
6.259
|
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like
|
|
chr12_-_16759430
|
6.240
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_+_41614911
|
6.234
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_+_49027770
|
6.233
|
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr3_+_183873215
|
6.219
|
|
DVL3
|
dishevelled, dsh homolog 3 (Drosophila)
|
|
chr1_+_1260142
|
6.218
|
NM_001029885
|
GLTPD1
|
glycolipid transfer protein domain containing 1
|
|
chr15_-_75230354
|
6.203
|
|
COX5A
|
cytochrome c oxidase subunit Va
|
|
chr20_+_32077904
|
6.202
|
NM_001032999
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr20_+_24450269
|
6.202
|
|
SYNDIG1
|
synapse differentiation inducing 1
|
|
chr20_-_44455932
|
6.186
|
NM_003279
|
TNNC2
|
troponin C type 2 (fast)
|
|
chr1_+_15764934
|
6.186
|
NM_007272
|
CTRC
|
chymotrypsin C (caldecrin)
|
|
chr2_+_219646704
|
6.177
|
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chr12_-_123466195
|
6.142
|
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr9_-_131872782
|
6.131
|
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr11_+_63742014
|
6.114
|
NM_004074
|
COX8A
|
cytochrome c oxidase subunit VIIIA (ubiquitous)
|
|
chr15_+_43885251
|
6.108
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chrX_+_152954965
|
6.083
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr3_+_183873305
|
6.062
|
|
DVL3
|
dishevelled, dsh homolog 3 (Drosophila)
|
|
chr1_-_154155698
|
6.057
|
NM_001043351
NM_001043352
NM_001043353
NM_153649
|
TPM3
|
tropomyosin 3
|
|
chr6_-_52860076
|
6.047
|
NM_001512
|
GSTA4
|
glutathione S-transferase alpha 4
|
|
chr1_-_144997110
|
6.037
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr11_-_119293848
|
6.023
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr2_-_97303983
|
5.997
|
|
KANSL3
|
KAT8 regulatory NSL complex subunit 3
|
|
chr6_+_27833033
|
5.979
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr1_+_152635871
|
5.954
|
NM_178430
|
LCE2D
|
late cornified envelope 2D
|
|
chr9_+_139872014
|
5.948
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr12_-_11036815
|
5.933
|
NM_006250
|
PRH1
|
proline-rich protein HaeIII subfamily 1
|
|
chr6_+_43738757
|
5.925
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr1_+_206224282
|
5.908
|
NM_000707
|
AVPR1B
|
arginine vasopressin receptor 1B
|
|
chr15_+_84115979
|
5.887
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr3_-_49460110
|
5.887
|
NM_000481
NM_001164710
NM_001164711
NM_001164712
|
AMT
|
aminomethyltransferase
|
|
chr10_-_81320141
|
5.881
|
NM_001098668
|
SFTPA1
SFTPA2
|
surfactant protein A1
surfactant protein A2
|
|
chr12_-_6233684
|
5.844
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr19_+_2328679
|
5.828
|
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr5_-_148034089
|
5.790
|
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr19_+_2328628
|
5.775
|
NM_001077238
NM_152988
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr15_+_43985083
|
5.769
|
NM_001015001
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chr1_-_149889362
|
5.757
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr11_+_45907046
|
5.732
|
NM_005456
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr1_-_149858114
|
5.670
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr6_+_31982538
|
5.668
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr19_-_39322411
|
5.666
|
NM_001398
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal
|
|
chr20_+_44036940
|
5.659
|
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr11_-_8832189
|
5.658
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr8_-_18541441
|
5.630
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr12_+_121163570
|
5.599
|
NM_000017
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain
|