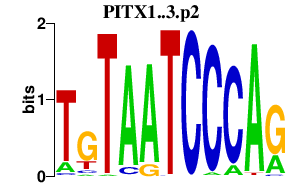
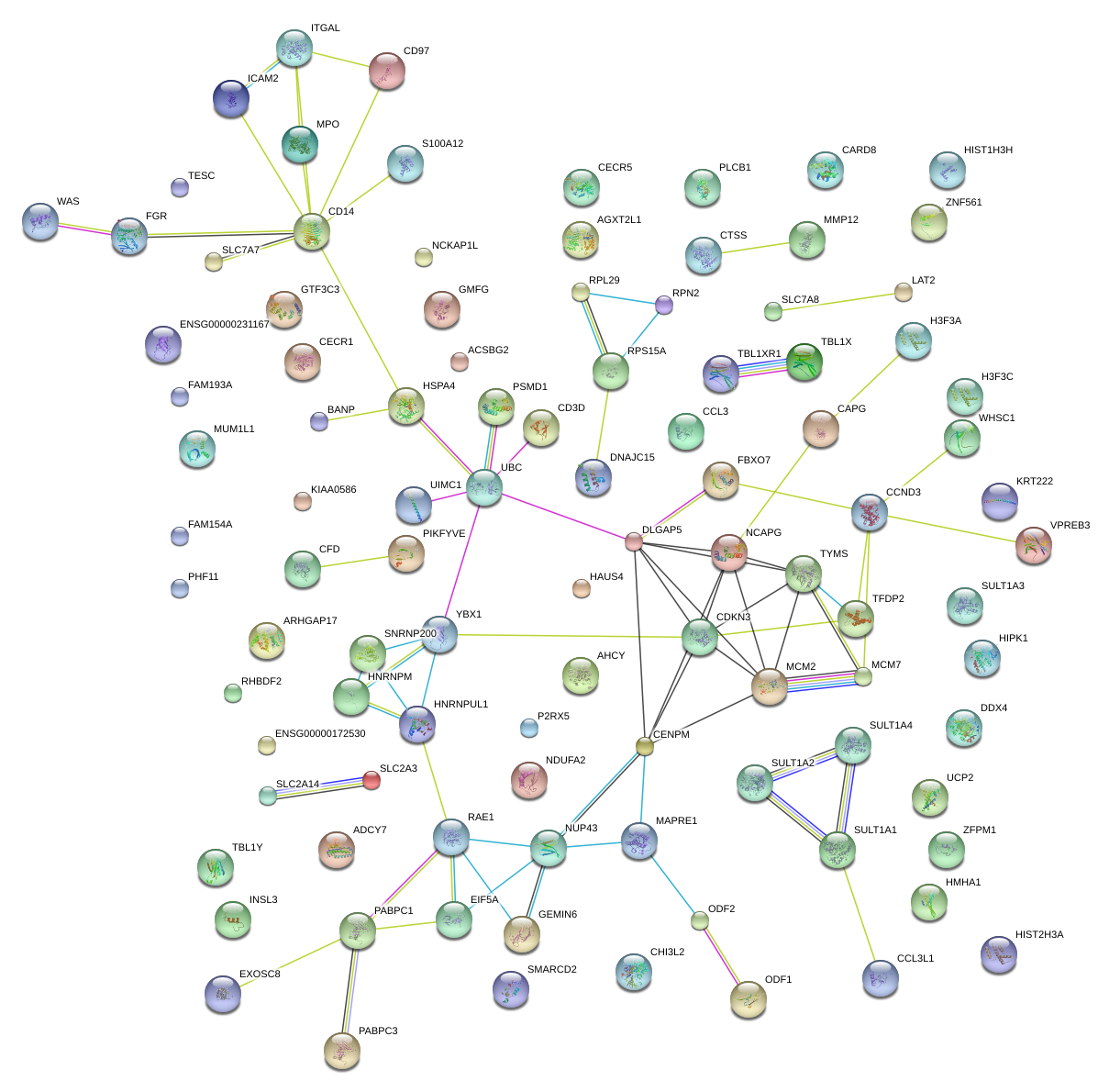
|
chr16_-_18798077
|
24.610
|
|
|
|
|
chr22_-_17680485
|
23.483
|
NM_177405
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr19_-_39826625
|
23.244
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr22_-_17690778
|
22.095
|
NM_017424
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr19_-_39826676
|
22.033
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr17_-_38821286
|
21.477
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr3_+_127317271
|
17.961
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr1_-_150738292
|
17.908
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr3_+_127317274
|
17.802
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr9_+_131217433
|
16.884
|
NM_153436
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr3_-_141719188
|
16.801
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr3_+_127317212
|
16.294
|
NM_004526
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr1_-_150738255
|
16.143
|
|
CTSS
|
cathepsin S
|
|
chr16_+_29471206
|
16.040
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr7_+_73624328
|
15.768
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr20_+_55926144
|
15.722
|
NM_003610
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr7_+_73624347
|
15.603
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr22_-_42336222
|
14.995
|
NM_001110215
|
CENPM
|
centromere protein M
|
|
chr12_-_117537090
|
14.832
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chr16_+_30210548
|
14.711
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr11_-_118213321
|
14.413
|
NM_000732
NM_001040651
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex)
|
|
chr1_+_114496498
|
14.301
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr16_+_50321822
|
14.271
|
NM_001114
|
ADCY7
|
adenylate cyclase 7
|
|
chr20_+_55926299
|
13.950
|
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr20_+_35807522
|
13.837
|
|
RPN2
|
ribophorin II
|
|
chr12_+_54892567
|
13.828
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr7_+_73624086
|
13.606
|
NM_014146
NM_032463
NM_032464
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr19_-_48744276
|
13.165
|
NM_001184900
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr19_+_8509594
|
13.151
|
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr1_-_153348057
|
13.073
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr19_-_10291056
|
12.963
|
|
|
|
|
chr16_+_88003623
|
12.748
|
NM_001173543
|
BANP
|
BTG3 associated nuclear protein
|
|
chr19_-_9731883
|
12.741
|
NM_152289
|
ZNF561
|
zinc finger protein 561
|
|
chr19_+_1071208
|
12.678
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr2_-_96950421
|
12.429
|
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5)
|
|
chr11_-_73693883
|
12.185
|
NM_003355
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier)
|
|
chr20_+_35807430
|
12.168
|
NM_001135771
NM_002951
|
RPN2
|
ribophorin II
|
|
chr19_+_14491920
|
12.114
|
NM_001025160
NM_001784
NM_078481
|
CD97
|
CD97 molecule
|
|
chr7_-_99699426
|
12.002
|
NM_005916
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr14_-_23285011
|
11.991
|
NM_001126105
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr1_-_150738232
|
11.841
|
|
CTSS
|
cathepsin S
|
|
chr12_-_8043743
|
11.764
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr19_-_17932312
|
11.588
|
NM_005543
|
INSL3
|
insulin-like 3 (Leydig cell)
|
|
chr13_+_50069745
|
11.530
|
NM_001040443
|
PHF11
|
PHD finger protein 11
|
|
chr16_+_30483959
|
11.405
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr17_-_56358266
|
11.243
|
NM_000250
|
MPO
|
myeloperoxidase
|
|
chr4_+_2627158
|
11.204
|
NM_003704
|
FAM193A
|
family with sequence similarity 193, member A
|
|
chr16_-_18801624
|
11.014
|
NM_001019
NM_001030009
|
RPS15A
|
ribosomal protein S15a
|
|
chr19_+_41768368
|
10.830
|
NM_144732
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr5_+_132406080
|
10.723
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr16_+_30484049
|
10.696
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr17_-_34625628
|
10.583
|
NM_001001437
NM_021006
|
CCL3L3
CCL3
CCL3L1
|
chemokine (C-C motif) ligand 3-like 3
chemokine (C-C motif) ligand 3
chemokine (C-C motif) ligand 3-like 1
|
|
chr20_-_32891075
|
10.302
|
NM_000687
|
AHCY
|
adenosylhomocysteinase
|
|
chr17_-_74483806
|
10.173
|
NM_001005498
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr4_+_1894508
|
10.136
|
NM_007331
NM_133330
NM_133331
NM_133335
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr20_+_31407696
|
10.115
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr11_-_73693820
|
9.938
|
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier)
|
|
chr19_+_6135650
|
9.876
|
NM_030924
|
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2
|
|
chr19_+_860615
|
9.860
|
|
CFD
|
complement factor D (adipsin)
|
|
chr6_+_31598366
|
9.813
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr13_+_37574677
|
9.809
|
NM_181503
|
EXOSC8
|
exosome component 8
|
|
chr2_+_39005326
|
9.804
|
NM_024775
|
GEMIN6
|
gem (nuclear organelle) associated protein 6
|
|
chr17_-_34524054
|
9.602
|
NM_001001437
NM_021006
|
CCL3L3
CCL3
CCL3L1
|
chemokine (C-C motif) ligand 3-like 3
chemokine (C-C motif) ligand 3
chemokine (C-C motif) ligand 3-like 1
|
|
chr22_+_32870699
|
9.374
|
NM_012179
|
FBXO7
|
F-box protein 7
|
|
chr22_-_17646176
|
9.370
|
NM_017829
|
CECR5
|
cat eye syndrome chromosome region, candidate 5
|
|
chr6_-_42016609
|
9.346
|
NM_001136017
NM_001136126
|
CCND3
|
cyclin D3
|
|
chr17_-_3599487
|
9.317
|
NM_001204519
NM_001204520
NM_002561
NM_175080
|
P2RX5-TAX1BP3
P2RX5
|
P2RX5-TAX1BP3 readthrough
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr5_+_55062712
|
9.304
|
NM_001166534
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr14_+_73957608
|
9.245
|
NM_024644
|
C14orf169
|
chromosome 14 open reading frame 169
|
|
chr13_+_50070096
|
9.171
|
|
PHF11
|
PHD finger protein 11
|
|
chr20_+_55926585
|
9.117
|
NM_001015885
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr22_-_42343079
|
9.027
|
NM_001002876
NM_024053
|
CENPM
|
centromere protein M
|
|
chr14_+_54863672
|
9.021
|
NM_001130851
NM_005192
|
CDKN3
|
cyclin-dependent kinase inhibitor 3
|
|
chr1_+_111772332
|
8.991
|
NM_001025199
|
CHI3L2
|
chitinase 3-like 2
|
|
chr12_+_54891539
|
8.977
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr16_-_25026650
|
8.871
|
NM_001006634
NM_018054
|
ARHGAP17
|
Rho GTPase activating protein 17
|
|
chr2_-_197664335
|
8.858
|
NM_001206774
NM_012086
|
GTF3C3
|
general transcription factor IIIC, polypeptide 3, 102kDa
|
|
chr14_+_58906398
|
8.820
|
NM_001244193
|
KIAA0586
|
KIAA0586
|
|
chr6_-_150057659
|
8.808
|
|
NUP43
|
nucleoporin 43kDa
|
|
chr12_+_54891522
|
8.748
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr22_-_24096551
|
8.650
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chrX_+_9431314
|
8.649
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_+_43162386
|
8.629
|
|
YBX1
|
Y box binding protein 1
|
|
chr12_+_54891491
|
8.523
|
NM_005337
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr12_-_46357949
|
8.503
|
|
|
|
|
chr3_-_52029957
|
8.490
|
NM_000992
|
RPL29
|
ribosomal protein L29
|
|
chr12_-_31945174
|
8.369
|
NM_001013699
|
H3F3C
|
H3 histone, family 3C
|
|
chr5_-_176433365
|
8.311
|
NM_016290
NM_001199297
NM_001199298
|
UIMC1
|
ubiquitin interaction motif containing 1
|
|
chr14_-_55650348
|
8.267
|
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chrX_+_48542159
|
8.110
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr6_-_31619481
|
8.092
|
|
|
|
|
chr14_-_23426356
|
8.061
|
|
HAUS4
|
HAUS augmin-like complex, subunit 4
|
|
chr17_-_61920256
|
8.011
|
|
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr5_-_140013285
|
7.994
|
NM_001040021
NM_001174104
NM_001174105
|
CD14
|
CD14 molecule
|
|
chr2_-_85637486
|
7.948
|
NM_001747
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr18_+_657674
|
7.861
|
|
TYMS
|
thymidylate synthetase
|
|
chr13_+_43597338
|
7.825
|
NM_013238
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15
|
|
chr17_+_7210317
|
7.793
|
NM_001143760
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr13_+_37574916
|
7.772
|
|
EXOSC8
|
exosome component 8
|
|
chr17_-_62097937
|
7.741
|
NM_001099786
NM_001099787
NM_001099788
NM_001099789
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr1_-_27952591
|
7.701
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr13_+_25670275
|
7.670
|
NM_030979
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3
|
|
chr11_+_68367824
|
7.649
|
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr5_+_132432880
|
7.609
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr14_-_23426265
|
7.544
|
NM_001166269
NM_001166270
NM_017815
|
HAUS4
|
HAUS augmin-like complex, subunit 4
|
|
chr11_-_615920
|
7.522
|
|
IRF7
|
interferon regulatory factor 7
|
|
chr6_+_34204928
|
7.479
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr17_+_41158741
|
7.420
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chr18_+_657633
|
7.410
|
|
TYMS
|
thymidylate synthetase
|
|
chr20_+_35807695
|
7.401
|
|
RPN2
|
ribophorin II
|
|
chr11_+_308106
|
7.386
|
NM_006435
|
IFITM2
|
interferon induced transmembrane protein 2 (1-8D)
|
|
chr2_+_68961912
|
7.381
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr11_-_67205151
|
7.345
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chr7_+_66386228
|
7.312
|
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr5_+_172386438
|
7.246
|
NM_016093
|
RPL26L1
|
ribosomal protein L26-like 1
|
|
chr19_+_3136026
|
7.228
|
NM_002068
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class)
|
|
chrX_-_129272014
|
7.087
|
NM_001130846
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr19_-_47197162
|
7.021
|
|
PRKD2
|
protein kinase D2
|
|
chr7_+_66386237
|
7.018
|
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr17_+_76210329
|
7.016
|
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr17_-_61920328
|
6.992
|
NM_001098426
|
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr6_-_32920812
|
6.986
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr20_-_44006876
|
6.974
|
NM_014477
|
TP53TG5
|
TP53 target 5
|
|
chr11_+_65101345
|
6.973
|
|
DPF2
|
D4, zinc and double PHD fingers family 2
|
|
chr11_+_17332454
|
6.971
|
|
|
|
|
chr6_-_31704272
|
6.955
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr19_+_17622445
|
6.936
|
|
PGLS
|
6-phosphogluconolactonase
|
|
chr19_+_48828627
|
6.934
|
NM_001425
|
EMP3
|
epithelial membrane protein 3
|
|
chr17_+_18768781
|
6.930
|
NM_001243940
|
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2
|
|
chr6_-_31774725
|
6.916
|
NM_021177
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr6_-_31704281
|
6.914
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr19_-_13213513
|
6.865
|
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr11_+_118955569
|
6.854
|
NM_000190
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr19_+_6361558
|
6.833
|
|
CLPP
|
ClpP caseinolytic peptidase, ATP-dependent, proteolytic subunit homolog (E. coli)
|
|
chr18_+_657461
|
6.799
|
NM_001071
|
TYMS
|
thymidylate synthetase
|
|
chr6_-_31704287
|
6.727
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr12_-_54689536
|
6.719
|
NM_006163
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr1_-_40042415
|
6.682
|
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr17_+_73201528
|
6.644
|
NM_024844
|
NUP85
|
nucleoporin 85kDa
|
|
chr7_+_66386129
|
6.600
|
NM_017994
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr6_-_31763551
|
6.588
|
NM_006295
|
VARS
|
valyl-tRNA synthetase
|
|
chr7_+_99070428
|
6.587
|
|
ZNF789
|
zinc finger protein 789
|
|
chr11_-_615998
|
6.575
|
NM_001572
NM_004029
|
IRF7
|
interferon regulatory factor 7
|
|
chr18_+_3453771
|
6.557
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr11_+_65101224
|
6.554
|
NM_006268
|
DPF2
|
D4, zinc and double PHD fingers family 2
|
|
chr1_-_40042461
|
6.545
|
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr13_+_103459495
|
6.507
|
NM_001204425
|
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough
basic, immunoglobulin-like variable motif containing
|
|
chr19_-_8642309
|
6.507
|
NM_012335
|
MYO1F
|
myosin IF
|
|
chr1_+_28832454
|
6.484
|
NM_001048199
|
RCC1
|
regulator of chromosome condensation 1
|
|
chr3_+_9476506
|
6.446
|
|
|
|
|
chr6_-_30640781
|
6.437
|
NM_001164239
NM_003587
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16
|
|
chr6_-_31704304
|
6.414
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr19_+_17622420
|
6.404
|
NM_012088
|
PGLS
|
6-phosphogluconolactonase
|
|
chr19_-_54876671
|
6.401
|
NM_002287
NM_021706
|
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1
|
|
chr17_-_30228715
|
6.354
|
NM_018428
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast)
|
|
chr7_+_5253816
|
6.347
|
NM_001033520
|
WIPI2
|
WD repeat domain, phosphoinositide interacting 2
|
|
chr6_-_31704318
|
6.332
|
NM_001288
|
CLIC1
|
chloride intracellular channel 1
|
|
chr2_-_70520845
|
6.288
|
NM_003096
|
SNRPG
|
small nuclear ribonucleoprotein polypeptide G
|
|
chr1_-_40042519
|
6.279
|
NM_001135653
NM_001135654
NM_003819
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr1_+_247581350
|
6.247
|
NM_001127462
NM_001243133
NM_004895
NM_183395
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chr1_-_225616556
|
6.241
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr3_-_126327397
|
6.230
|
NM_001039783
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor
|
|
chr3_-_50375650
|
6.228
|
NM_170712
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr6_-_31510251
|
6.225
|
NM_004640
NM_080598
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B
|
|
chr6_+_139456240
|
6.205
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr5_+_118691549
|
6.204
|
NM_014350
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr15_+_29077439
|
6.183
|
|
|
|
|
chr9_+_132565394
|
6.128
|
NM_014506
|
TOR1B
|
torsin family 1, member B (torsin B)
|
|
chr2_+_87140905
|
6.115
|
NM_001078170
|
RGPD2
|
RANBP2-like and GRIP domain containing 2
|
|
chr17_-_3599359
|
6.115
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr7_-_44614136
|
6.079
|
NM_019082
|
DDX56
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 56
|
|
chr22_-_42611310
|
6.057
|
NM_005650
NM_181492
|
TCF20
|
transcription factor 20 (AR1)
|
|
chr1_-_40349105
|
6.053
|
|
TRIT1
|
tRNA isopentenyltransferase 1
|
|
chr19_+_7504573
|
6.053
|
NM_001130955
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr17_+_48821106
|
5.999
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr6_-_30640618
|
5.984
|
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16
|
|
chr19_-_22966972
|
5.956
|
NM_001080409
|
ZNF99
|
zinc finger protein 99
|
|
chr1_-_1711453
|
5.939
|
NM_001198994
|
NADK
|
NAD kinase
|
|
chr12_-_13248581
|
5.923
|
NM_001206843
NM_001206845
NM_031289
NM_153823
|
GSG1
|
germ cell associated 1
|
|
chr5_+_118691692
|
5.918
|
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr11_+_60197061
|
5.900
|
NM_023945
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5
|
|
chr19_-_8642297
|
5.886
|
|
MYO1F
|
myosin IF
|
|
chr1_+_236681513
|
5.868
|
NM_201545
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr1_+_161185061
|
5.859
|
|
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide
|
|
chr20_+_361272
|
5.858
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr19_-_8642337
|
5.819
|
|
MYO1F
|
myosin IF
|
|
chr12_-_120638912
|
5.810
|
NM_001002
NM_053275
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr12_-_123706393
|
5.807
|
NM_022782
|
MPHOSPH9
|
M-phase phosphoprotein 9
|
|
chr16_+_30087342
|
5.786
|
NM_002720
|
PPP4C
|
protein phosphatase 4, catalytic subunit
|
|
chr16_+_28943259
|
5.780
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr2_-_27357519
|
5.757
|
NM_013388
|
PREB
|
prolactin regulatory element binding
|
|
chr1_+_26872306
|
5.753
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr14_-_106452693
|
5.721
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chrX_+_49028183
|
5.712
|
NM_002668
|
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched)
|
|
chr11_-_64647136
|
5.663
|
|
EHD1
|
EH-domain containing 1
|
|
chr3_-_128369525
|
5.640
|
|
RPN1
|
ribophorin I
|
|
chr19_+_36024313
|
5.599
|
NM_014364
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic
|
|
chr7_+_99955625
|
5.593
|
NM_178238
|
PILRB
|
paired immunoglobin-like type 2 receptor beta
|
|
chr12_+_57881853
|
5.569
|
|
MARS
|
methionyl-tRNA synthetase
|
|
chr17_-_56358161
|
5.566
|
|
MPO
|
myeloperoxidase
|