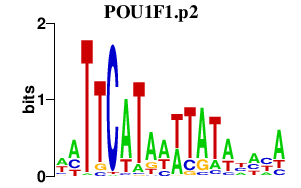
|
chr1_-_93399531
|
49.766
|
|
|
|
|
chr12_-_91573264
|
45.112
|
NM_133503
|
DCN
|
decorin
|
|
chr6_-_52628272
|
34.139
|
NM_000846
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr12_-_45307710
|
31.768
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr4_+_74270832
|
30.682
|
|
ALB
|
albumin
|
|
chr14_-_107013474
|
29.248
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_+_196857143
|
27.912
|
NM_001201550
NM_001201551
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr4_+_106631966
|
27.707
|
NM_001031720
|
GSTCD
|
glutathione S-transferase, C-terminal domain containing
|
|
chr6_-_33041266
|
27.577
|
NM_033554
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr7_-_64023444
|
27.345
|
|
|
|
|
chr1_+_196743929
|
25.784
|
NM_001166624
NM_021023
|
CFHR3
|
complement factor H-related 3
|
|
chr5_-_41213513
|
25.269
|
NM_000065
|
C6
|
complement component 6
|
|
chr12_+_21679190
|
25.163
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr1_-_163172863
|
24.723
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_-_161279724
|
23.629
|
NM_000530
|
MPZ
|
myelin protein zero
|
|
chr8_-_86253863
|
23.282
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|
|
chr12_-_87232635
|
22.954
|
NM_013244
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative)
|
|
chr4_+_71337833
|
22.892
|
NM_152291
|
MUC7
|
mucin 7, secreted
|
|
chr4_+_71337674
|
22.428
|
NM_001145007
|
MUC7
|
mucin 7, secreted
|
|
chr16_-_5116030
|
22.050
|
NM_001098514
NM_152459
|
C16orf89
|
chromosome 16 open reading frame 89
|
|
chr5_+_40909617
|
21.966
|
|
C7
|
complement component 7
|
|
chr5_+_40909558
|
21.616
|
NM_000587
|
C7
|
complement component 7
|
|
chr6_+_72922513
|
21.526
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr10_+_104613966
|
21.297
|
NM_001136200
NM_144591
|
C10orf32-AS3MT
C10orf32
|
C10orf32-AS3MT readthrough
chromosome 10 open reading frame 32
|
|
chr20_+_30102261
|
21.042
|
|
HM13
|
histocompatibility (minor) 13
|
|
chr3_-_121379757
|
20.854
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr4_-_100242487
|
20.355
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr3_-_121379701
|
19.631
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr14_-_106610338
|
19.607
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr5_-_75919051
|
19.555
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr4_+_71263598
|
19.337
|
NM_021225
|
PROL1
|
proline rich, lacrimal 1
|
|
chr2_-_26700909
|
19.247
|
NM_004802
NM_194322
NM_194323
|
OTOF
|
otoferlin
|
|
chr6_+_153019029
|
18.916
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr18_-_40695483
|
18.890
|
NM_002930
|
RIT2
|
Ras-like without CAAX 2
|
|
chr12_-_91576579
|
18.756
|
|
DCN
|
decorin
|
|
chr3_+_122296432
|
18.673
|
NM_001113523
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr10_+_101088794
|
18.268
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chrX_+_7810302
|
18.200
|
NM_013452
|
VCX
|
variable charge, X-linked
|
|
chr6_-_22297729
|
18.015
|
NM_000948
|
PRL
|
prolactin
|
|
chr5_+_54320083
|
17.988
|
NM_002104
|
GZMK
|
granzyme K (granzyme 3; tryptase II)
|
|
chr11_-_114466397
|
17.926
|
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr9_-_21368005
|
17.263
|
NM_006900
|
IFNA1
IFNA13
|
interferon, alpha 1
interferon, alpha 13
|
|
chr1_+_196788860
|
17.239
|
NM_002113
|
CFHR1
|
complement factor H-related 1
|
|
chr15_-_45670624
|
16.775
|
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr17_-_29641068
|
16.589
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr3_+_38347426
|
16.541
|
NM_004803
|
SLC22A14
|
solute carrier family 22, member 14
|
|
chr4_-_71532341
|
16.124
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr19_+_42381189
|
15.922
|
NM_001783
NM_021601
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha
|
|
chr6_+_32407618
|
15.876
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chrY_-_6742028
|
15.875
|
NM_001143
|
AMELY
|
amelogenin, Y-linked
|
|
chr22_+_24033047
|
15.841
|
NM_153615
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4
|
|
chr14_-_106774311
|
15.511
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr1_+_196912910
|
15.421
|
NM_005666
|
CFHR2
|
complement factor H-related 2
|
|
chr16_-_21314371
|
15.400
|
NM_001014444
NM_001888
|
CRYM
|
crystallin, mu
|
|
chr4_+_74269971
|
15.379
|
NM_000477
|
ALB
|
albumin
|
|
chr6_+_28109611
|
15.351
|
NM_006298
|
ZNF192
|
zinc finger protein 192
|
|
chr4_-_100273831
|
15.299
|
NM_000669
|
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide
|
|
chr18_-_64271215
|
15.231
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr6_-_32634287
|
15.110
|
NM_001243961
NM_002123
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr1_-_57045214
|
14.991
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_+_79347487
|
14.935
|
NM_002909
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr9_-_21187529
|
14.914
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr11_-_114466471
|
14.597
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr21_-_31538970
|
14.582
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr3_+_108021331
|
14.518
|
NM_007072
|
HHLA2
|
HERV-H LTR-associating 2
|
|
chr5_-_58295758
|
14.369
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_172389827
|
14.068
|
NM_139240
|
C1orf105
|
chromosome 1 open reading frame 105
|
|
chrX_-_2882288
|
13.921
|
NM_000047
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr1_+_147374945
|
13.882
|
NM_005267
|
GJA8
|
gap junction protein, alpha 8, 50kDa
|
|
chr8_-_82395401
|
13.825
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr1_+_153003677
|
13.595
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr17_-_34313685
|
13.440
|
NM_032962
NM_032963
|
CCL14
|
chemokine (C-C motif) ligand 14
|
|
chrY_+_20297334
|
13.310
|
NM_001002906
NM_004677
|
XKRY2
XKRY
|
XK, Kell blood group complex subunit-related, Y-linked 2
XK, Kell blood group complex subunit-related, Y-linked
|
|
chr6_+_131958441
|
13.086
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr12_+_7167979
|
13.047
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr14_-_101036130
|
13.018
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr1_-_35497563
|
13.008
|
NM_007167
|
ZMYM6
|
zinc finger, MYM-type 6
|
|
chr12_+_57157091
|
12.979
|
NM_003725
|
HSD17B6
|
hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse)
|
|
chr4_-_71532206
|
12.857
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr16_-_3627353
|
12.811
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr14_-_64749530
|
12.791
|
NM_001214902
NM_001214903
|
ESR2
|
estrogen receptor 2 (ER beta)
|
|
chr3_-_195310985
|
12.762
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr9_+_21440452
|
12.598
|
NM_024013
|
IFNA1
|
interferon, alpha 1
|
|
chr19_+_39971051
|
12.570
|
NM_001001563
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae)
|
|
chr4_+_88720701
|
12.479
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr7_+_102023016
|
12.461
|
|
|
|
|
chr19_-_21646685
|
12.360
|
|
|
|
|
chr2_-_77749335
|
12.127
|
NM_001134745
NM_024993
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4
|
|
chr9_-_23821842
|
12.121
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr4_+_88742549
|
12.110
|
NM_001184694
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr6_-_52668599
|
12.087
|
NM_145740
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chr11_+_124824076
|
12.034
|
|
CCDC15
|
coiled-coil domain containing 15
|
|
chrX_+_66788682
|
11.827
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr9_-_70429730
|
11.731
|
NM_001099279
NM_199244
|
FOXD4L2
FOXD4L4
|
forkhead box D4-like 2
forkhead box D4-like 4
|
|
chr5_-_39203061
|
11.614
|
|
FYB
|
FYN binding protein
|
|
chr9_-_21350885
|
11.594
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr20_+_30102237
|
11.573
|
NM_030789
NM_178580
NM_178581
NM_178582
|
HM13
|
histocompatibility (minor) 13
|
|
chr1_+_160336856
|
11.363
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr4_-_100212085
|
11.150
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr9_-_123812535
|
11.062
|
NM_001735
|
C5
|
complement component 5
|
|
chr19_+_840970
|
11.048
|
NM_002777
|
PRTN3
|
proteinase 3
|
|
chr11_-_36619776
|
10.995
|
NM_000536
NM_001243785
NM_001243786
|
RAG2
|
recombination activating gene 2
|
|
chr1_+_207277577
|
10.860
|
NM_000715
|
C4BPA
|
complement component 4 binding protein, alpha
|
|
chrY_-_19882311
|
10.764
|
NM_001002906
NM_004677
|
XKRY2
XKRY
|
XK, Kell blood group complex subunit-related, Y-linked 2
XK, Kell blood group complex subunit-related, Y-linked
|
|
chr6_+_72926462
|
10.663
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr16_+_67381252
|
10.601
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr13_+_111855406
|
10.598
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr9_+_102677457
|
10.579
|
|
STX17
|
syntaxin 17
|
|
chr3_+_148447966
|
10.571
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr4_+_69681709
|
10.565
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr17_+_33448597
|
10.466
|
NM_017559
|
FNDC8
|
fibronectin type III domain containing 8
|
|
chr6_-_32920812
|
10.460
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chrX_-_138724851
|
10.344
|
NM_001171877
NM_001171878
NM_001171879
NM_005369
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr6_-_49844808
|
10.289
|
NM_001205220
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr1_-_217113007
|
10.242
|
NM_001243512
NM_001243513
NM_001243510
NM_001243511
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr4_+_71226477
|
10.205
|
NM_012390
|
SMR3B
SMR3A
|
submaxillary gland androgen regulated protein 3B
submaxillary gland androgen regulated protein 3A
|
|
chr9_-_23821808
|
10.117
|
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr7_+_37723378
|
9.916
|
|
|
|
|
chr9_+_42717233
|
9.884
|
NM_001099279
NM_199244
|
FOXD4L2
FOXD4L4
|
forkhead box D4-like 2
forkhead box D4-like 4
|
|
chr2_+_79347610
|
9.880
|
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr7_-_14880954
|
9.690
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr12_-_52886971
|
9.686
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr10_+_32856650
|
9.578
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr2_-_174767282
|
9.564
|
|
|
|
|
chr3_+_101546833
|
9.534
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr12_-_371994
|
9.469
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr4_-_87028805
|
9.447
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr16_-_23724820
|
9.444
|
NM_033266
|
ERN2
|
endoplasmic reticulum to nucleus signaling 2
|
|
chr1_+_171154387
|
9.341
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr8_-_42623928
|
9.231
|
NM_001199279
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr12_-_10282680
|
9.145
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr9_-_70178814
|
9.124
|
NM_001126334
|
FOXD4L5
|
forkhead box D4-like 5
|
|
chr1_+_92632608
|
9.109
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr11_-_128457452
|
9.102
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr7_+_123565908
|
8.928
|
NM_001174045
NM_001174046
|
SPAM1
|
sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding)
|
|
chr9_-_21142143
|
8.764
|
NM_002177
|
IFNW1
|
interferon, omega 1
|
|
chr14_-_106405633
|
8.760
|
|
IGHM
|
immunoglobulin heavy constant mu
|
|
chr8_+_24151579
|
8.749
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr4_-_47983430
|
8.722
|
NM_001142564
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr1_+_152881022
|
8.704
|
NM_005547
|
IVL
|
involucrin
|
|
chr4_+_187148624
|
8.662
|
NM_000892
|
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1
|
|
chr12_-_10151689
|
8.653
|
NM_001099431
NM_016509
|
CLEC1B
|
C-type lectin domain family 1, member B
|
|
chr3_-_194072004
|
8.594
|
NM_001080513
|
CPN2
|
carboxypeptidase N, polypeptide 2
|
|
chr13_-_46756295
|
8.592
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_114522029
|
8.547
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr13_+_49684388
|
8.517
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr12_-_30830143
|
8.444
|
NM_001190995
|
IPO8
|
importin 8
|
|
chr11_-_85376159
|
8.427
|
NM_001039618
|
CREBZF
|
CREB/ATF bZIP transcription factor
|
|
chr8_-_17752847
|
8.405
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr10_+_96443250
|
8.334
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr17_-_3301703
|
8.315
|
NM_003553
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1
|
|
chr1_-_20445991
|
8.289
|
NM_012400
|
PLA2G2D
|
phospholipase A2, group IID
|
|
chr4_-_39033981
|
8.282
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr3_+_46619075
|
8.137
|
NM_003212
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr4_+_70894129
|
8.133
|
NM_000200
|
HTN3
|
histatin 3
|
|
chr5_-_149792276
|
8.128
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr4_-_70505359
|
8.083
|
NM_001105677
|
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2
|
|
chr6_-_32557431
|
8.082
|
NM_001243965
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr4_-_76944585
|
8.023
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr9_+_100263461
|
8.013
|
NM_001166116
|
TMOD1
|
tropomodulin 1
|
|
chr14_-_106641585
|
7.967
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_-_5304579
|
7.938
|
NM_005059
NM_134441
|
RLN2
|
relaxin 2
|
|
chrX_+_119384490
|
7.900
|
NM_006777
NM_001184742
|
ZBTB33
|
zinc finger and BTB domain containing 33
|
|
chr9_+_71944240
|
7.899
|
NM_004816
|
FAM189A2
|
family with sequence similarity 189, member A2
|
|
chr4_+_74281884
|
7.882
|
|
ALB
|
albumin
|
|
chr1_-_13390747
|
7.866
|
NM_001012276
|
PRAMEF8
PRAMEF7
|
PRAME family member 8
PRAME family member 7
|
|
chrX_-_19988381
|
7.838
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr5_+_156696351
|
7.837
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr2_+_68592317
|
7.830
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr1_-_57045235
|
7.829
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr20_-_45061554
|
7.822
|
|
ELMO2
|
engulfment and cell motility 2
|
|
chr5_+_140220811
|
7.822
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr7_+_137761177
|
7.811
|
NM_001190906
NM_001190907
NM_005989
|
AKR1D1
|
aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase)
|
|
chr5_-_997408
|
7.798
|
NM_001242737
|
LOC100506688
|
uncharacterized LOC100506688
|
|
chr17_+_74463629
|
7.781
|
NM_001088
|
AANAT
|
aralkylamine N-acetyltransferase
|
|
chr19_-_48389514
|
7.700
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr11_-_102401430
|
7.664
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr7_+_29519485
|
7.636
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr8_-_87755880
|
7.604
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr4_+_71108304
|
7.579
|
NM_005212
|
CSN3
|
casein kappa
|
|
chr19_+_48949241
|
7.578
|
|
GRWD1
|
glutamate-rich WD repeat containing 1
|
|
chr19_+_840963
|
7.548
|
|
PRTN3
|
proteinase 3
|
|
chr12_+_56324945
|
7.492
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr3_+_151488214
|
7.491
|
|
LOC201651
|
arylacetamide deacetylase (esterase) pseudogene
|
|
chr4_+_113739203
|
7.490
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr4_-_100356449
|
7.486
|
NM_000673
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr4_-_16077740
|
7.481
|
NM_001145849
NM_001145850
NM_001145851
NM_001145852
NM_006017
|
PROM1
|
prominin 1
|
|
chr6_-_149806116
|
7.451
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr21_-_15918635
|
7.428
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr2_+_68592432
|
7.381
|
|
PLEK
|
pleckstrin
|
|
chr2_+_169921298
|
7.350
|
NM_005771
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr6_+_26087447
|
7.348
|
NM_000410
NM_139003
NM_139004
NM_139006
NM_139007
NM_139008
NM_139009
NM_139010
NM_139011
|
HFE
|
hemochromatosis
|
|
chr7_+_143412938
|
7.223
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr4_+_175839508
|
7.193
|
NM_001130703
NM_001130704
NM_001130705
NM_014269
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr12_-_21487831
|
7.186
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr5_-_115152322
|
7.142
|
NM_001801
|
CDO1
|
cysteine dioxygenase, type I
|
|
chr6_-_90024936
|
7.119
|
NM_002043
|
GABRR2
|
gamma-aminobutyric acid (GABA) receptor, rho 2
|
|
chr4_-_70080448
|
7.089
|
NM_001073
|
UGT2B11
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B11
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chrX_-_49965657
|
7.073
|
NM_003886
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr16_-_21663908
|
7.072
|
|
IGSF6
|
immunoglobulin superfamily, member 6
|