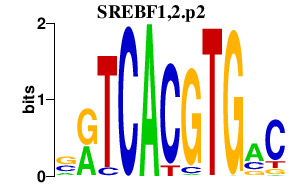
|
chr13_+_113951537
|
58.019
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr13_+_113951589
|
55.313
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr13_+_113951461
|
52.717
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr19_+_40854529
|
35.401
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr19_+_40854331
|
34.774
|
NM_001031696
NM_012268
|
PLD3
|
phospholipase D family, member 3
|
|
chr17_+_78075299
|
28.528
|
NM_000152
NM_001079803
NM_001079804
|
GAA
|
glucosidase, alpha; acid
|
|
chr17_+_78075390
|
27.735
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr20_+_44520021
|
27.387
|
|
CTSA
|
cathepsin A
|
|
chr8_-_54755836
|
25.560
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr8_-_54755821
|
25.532
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr20_+_44519965
|
24.564
|
|
CTSA
|
cathepsin A
|
|
chr8_-_54755546
|
23.497
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr19_-_48018163
|
23.428
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr15_-_72668301
|
23.213
|
NM_000520
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr1_+_44440595
|
22.851
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr16_-_67514989
|
22.478
|
NM_004691
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1
|
|
chr1_+_44440623
|
22.396
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr20_+_44519590
|
22.109
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr11_-_6640644
|
22.066
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr1_+_44440642
|
21.583
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr15_-_73925555
|
21.490
|
|
NPTN
|
neuroplastin
|
|
chr10_-_73610958
|
21.307
|
|
PSAP
|
prosaposin
|
|
chr10_-_73611005
|
20.695
|
|
PSAP
|
prosaposin
|
|
chr20_+_44519924
|
20.439
|
|
CTSA
|
cathepsin A
|
|
chr5_-_172198160
|
20.336
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr11_-_1785126
|
20.069
|
|
CTSD
|
cathepsin D
|
|
chr10_+_51827710
|
19.721
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr15_-_73925658
|
19.685
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr12_-_65153182
|
18.981
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr3_+_113465910
|
18.569
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr3_+_113465860
|
18.501
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr16_-_1525020
|
18.364
|
|
CLCN7
|
chloride channel 7
|
|
chr15_-_72668184
|
17.951
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr19_-_48018317
|
17.945
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr1_+_44440673
|
17.920
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr11_-_6640578
|
17.221
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr19_+_7587436
|
17.018
|
NM_020533
|
MCOLN1
|
mucolipin 1
|
|
chrX_+_101380659
|
16.948
|
NM_080390
|
TCEAL2
|
transcription elongation factor A (SII)-like 2
|
|
chr2_+_220042948
|
16.934
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr12_-_65153189
|
16.763
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr9_+_115913266
|
16.692
|
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr16_+_2570322
|
16.608
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr11_-_1785104
|
16.547
|
|
CTSD
|
cathepsin D
|
|
chr9_+_115913227
|
16.456
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr19_-_48018277
|
16.375
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr11_-_6640645
|
16.245
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr3_+_158519911
|
15.754
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr2_+_228189919
|
15.423
|
|
MFF
|
mitochondrial fission factor
|
|
chr5_-_150537261
|
15.319
|
|
ANXA6
|
annexin A6
|
|
chr2_+_220042986
|
15.250
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr6_-_33386064
|
15.216
|
NM_001014433
NM_001014837
NM_001014838
NM_001014840
NM_015921
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr3_+_158519888
|
14.670
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr19_-_10764449
|
14.621
|
|
LOC147727
|
uncharacterized LOC147727
|
|
chr11_-_1785087
|
14.521
|
|
CTSD
|
cathepsin D
|
|
chr5_-_150537306
|
14.502
|
|
ANXA6
|
annexin A6
|
|
chr20_-_39317875
|
14.433
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr1_-_154531083
|
13.920
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr11_+_121322848
|
13.911
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr20_+_44520203
|
13.868
|
|
CTSA
|
cathepsin A
|
|
chr18_+_9708187
|
13.818
|
NM_006868
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr18_+_9708269
|
13.687
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chrX_+_102470003
|
13.660
|
NM_001080425
NM_001127688
|
BEX4
|
brain expressed, X-linked 4
|
|
chr3_+_158519714
|
13.635
|
NM_001167903
NM_022736
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr17_-_35969401
|
13.619
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr22_+_35695872
|
13.404
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr16_-_1525006
|
13.347
|
|
CLCN7
|
chloride channel 7
|
|
chr1_+_110162427
|
13.294
|
NM_139156
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr18_+_9708322
|
13.286
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr3_+_112280871
|
13.159
|
NM_017945
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr3_+_158519877
|
13.135
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr3_+_158519925
|
13.116
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chrX_+_100663229
|
13.047
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr12_-_65153148
|
12.927
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chrX_+_100663120
|
12.838
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr1_+_110162459
|
12.695
|
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr3_+_128445024
|
12.619
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr1_-_40562909
|
12.462
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr16_+_85061303
|
12.429
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr16_-_5083913
|
12.423
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr10_+_51827654
|
12.412
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr1_-_40562917
|
12.381
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr16_-_5083933
|
12.353
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr19_+_1067293
|
12.193
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr10_+_46222647
|
12.170
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
|
family with sequence similarity 21, member C
|
|
chr16_-_1525040
|
12.142
|
NM_001114331
NM_001287
|
CLCN7
|
chloride channel 7
|
|
chr18_-_29523087
|
12.092
|
NM_014939
|
TRAPPC8
|
trafficking protein particle complex 8
|
|
chr1_-_1167421
|
11.904
|
NM_016176
NM_016547
|
SDF4
|
stromal cell derived factor 4
|
|
chr8_-_17941598
|
11.889
|
|
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1
|
|
chr10_-_73611041
|
11.885
|
|
PSAP
|
prosaposin
|
|
chr1_-_154928564
|
11.795
|
NM_020524
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr8_-_11725509
|
11.396
|
|
CTSB
|
cathepsin B
|
|
chr11_+_118938473
|
11.212
|
NM_021729
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr1_-_241520384
|
11.205
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr2_-_197036262
|
11.161
|
|
STK17B
|
serine/threonine kinase 17b
|
|
chr16_-_88923233
|
10.854
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr1_-_241520467
|
10.819
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chrX_+_100663205
|
10.797
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr10_-_73611050
|
10.766
|
NM_001042465
NM_001042466
NM_002778
|
PSAP
|
prosaposin
|
|
chr4_-_15656944
|
10.735
|
|
FBXL5
|
F-box and leucine-rich repeat protein 5
|
|
chr11_-_85780088
|
10.656
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr3_+_112280911
|
10.620
|
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr17_-_76124767
|
10.526
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr11_+_118938528
|
10.502
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr12_+_12869818
|
10.382
|
NM_004064
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr8_-_17941614
|
10.301
|
|
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1
|
|
chr1_-_39325317
|
10.299
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr22_-_39268230
|
10.284
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr11_+_118938515
|
10.133
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr5_-_150537335
|
9.930
|
NM_001155
|
ANXA6
|
annexin A6
|
|
chr9_+_6757636
|
9.851
|
NM_001146694
NM_001146695
NM_015061
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr7_-_30029284
|
9.835
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr22_+_35695792
|
9.782
|
NM_001135730
NM_001135732
NM_005488
|
TOM1
|
target of myb1 (chicken)
|
|
chr20_-_36156226
|
9.665
|
|
BLCAP
|
bladder cancer associated protein
|
|
chr15_-_72668740
|
9.586
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr16_-_28503622
|
9.554
|
NM_001042432
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr11_+_118938530
|
9.516
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr16_-_15736964
|
9.439
|
NM_001184998
NM_001184999
NM_014647
|
KIAA0430
|
KIAA0430
|
|
chr1_+_11866240
|
9.428
|
|
CLCN6
|
chloride channel 6
|
|
chr3_+_113465901
|
9.393
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chrX_+_103031797
|
9.118
|
|
PLP1
|
proteolipid protein 1
|
|
chr22_-_21213069
|
9.104
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr22_-_21213099
|
8.985
|
NM_058004
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr16_+_87425946
|
8.985
|
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta
|
|
chr22_-_21213012
|
8.924
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr14_+_96505566
|
8.707
|
NM_001252507
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr11_-_6640661
|
8.640
|
NM_000391
|
TPP1
|
tripeptidyl peptidase I
|
|
chr20_-_4795768
|
8.631
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr20_-_36156282
|
8.619
|
NM_001167820
NM_006698
|
BLCAP
|
bladder cancer associated protein
|
|
chr8_-_11725622
|
8.580
|
|
CTSB
|
cathepsin B
|
|
chr13_-_50159706
|
8.553
|
NM_018191
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr2_+_220042901
|
8.516
|
NM_024293
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr15_+_89182038
|
8.503
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr17_+_42422627
|
8.480
|
|
GRN
|
granulin
|
|
chr16_+_19567021
|
8.469
|
|
C16orf62
|
chromosome 16 open reading frame 62
|
|
chr19_+_1067536
|
8.454
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr20_-_36156082
|
8.450
|
NM_001167822
NM_001167821
|
BLCAP
|
bladder cancer associated protein
|
|
chr12_+_51632620
|
8.423
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr9_+_117349993
|
8.290
|
NM_004888
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chr13_-_50159628
|
8.279
|
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chrX_-_15872921
|
8.275
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr12_-_65153055
|
8.263
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr12_+_51632654
|
8.216
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr1_-_154600383
|
8.167
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr6_+_160390128
|
8.107
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr19_+_50016500
|
8.078
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr3_-_4508939
|
8.062
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr3_+_128444978
|
8.035
|
NM_004637
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr6_-_33385662
|
8.018
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr16_+_2564000
|
7.993
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr19_+_50016490
|
7.948
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chrX_+_103031753
|
7.944
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr12_+_51632507
|
7.920
|
NM_001136264
NM_001136266
NM_001136267
NM_001136268
NM_001136269
NM_014764
|
DAZAP2
|
DAZ associated protein 2
|
|
chr12_+_112451151
|
7.913
|
NM_001034025
NM_006817
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chrX_-_15872882
|
7.908
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr16_+_87425998
|
7.904
|
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta
|
|
chr18_+_9708299
|
7.889
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr22_+_35777087
|
7.868
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr12_+_51632589
|
7.808
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr1_-_1167358
|
7.742
|
|
SDF4
|
stromal cell derived factor 4
|
|
chr16_+_87425874
|
7.695
|
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta
|
|
chr12_-_109027654
|
7.688
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr7_+_151722774
|
7.661
|
NM_022087
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chr20_+_57428035
|
7.644
|
NM_001077490
NM_080425
|
GNAS
|
GNAS complex locus
|
|
chr12_+_6833425
|
7.605
|
NM_001164093
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chrX_-_15872938
|
7.597
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr16_+_2563993
|
7.536
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr17_+_25621141
|
7.435
|
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr7_+_149570004
|
7.432
|
NM_001100592
NM_145230
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chrX_+_153656977
|
7.431
|
NM_001183
|
ATP6AP1
|
ATPase, H+ transporting, lysosomal accessory protein 1
|
|
chrX_-_15872905
|
7.421
|
|
|
|
|
chrX_+_153657025
|
7.415
|
|
ATP6AP1
|
ATPase, H+ transporting, lysosomal accessory protein 1
|
|
chr8_-_11725595
|
7.401
|
|
CTSB
|
cathepsin B
|
|
chr11_-_1785190
|
7.356
|
NM_001909
|
CTSD
|
cathepsin D
|
|
chr3_+_5021096
|
7.332
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr9_-_123476578
|
7.296
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr16_+_2563914
|
7.274
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr19_+_18208596
|
7.271
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr8_-_11725587
|
7.248
|
|
CTSB
|
cathepsin B
|
|
chr16_+_2563977
|
7.238
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chrX_+_153657119
|
7.236
|
|
ATP6AP1
|
ATPase, H+ transporting, lysosomal accessory protein 1
|
|
chr8_+_26240522
|
7.193
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr12_-_109027565
|
7.177
|
|
SELPLG
|
selectin P ligand
|
|
chr6_+_138188352
|
7.125
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr11_-_85780105
|
7.083
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr16_-_28503079
|
7.061
|
NM_000086
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr8_-_17941617
|
7.038
|
NM_177924
|
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1
|
|
chr16_-_88923348
|
6.972
|
NM_000512
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr1_-_154928595
|
6.970
|
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chrX_-_13835313
|
6.932
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr7_+_108210308
|
6.929
|
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr17_+_42422490
|
6.908
|
NM_002087
|
GRN
|
granulin
|
|
chr19_+_1067114
|
6.881
|
NM_012292
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr3_+_147127151
|
6.878
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr6_-_33385814
|
6.877
|
|
|
|
|
chr3_+_5021141
|
6.862
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr16_+_11762235
|
6.777
|
NM_003498
|
SNN
|
stannin
|
|
chr7_+_151722842
|
6.765
|
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chr13_+_42846255
|
6.669
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr11_+_65479472
|
6.663
|
NM_001206833
NM_006388
NM_182709
NM_182710
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr14_-_74551159
|
6.647
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|