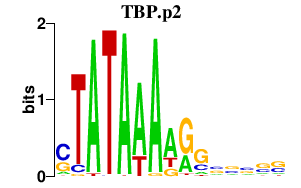
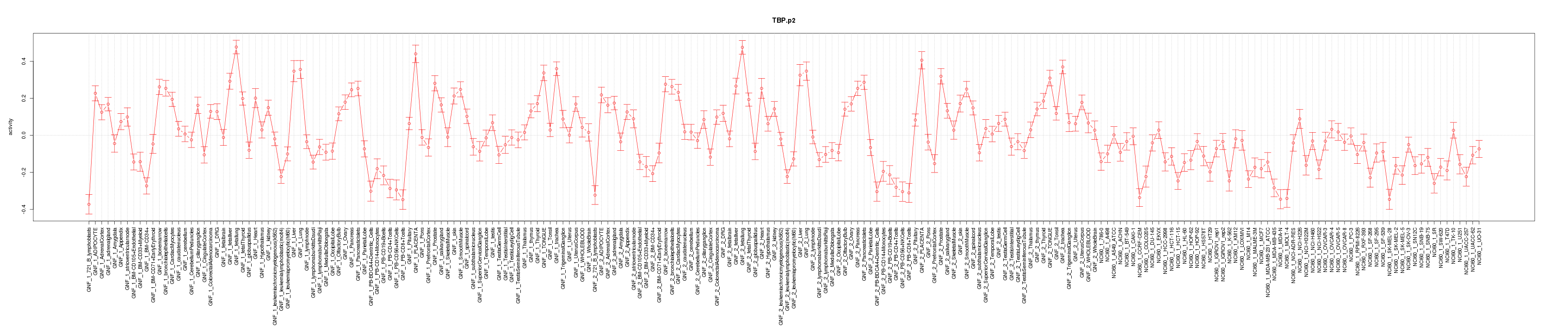
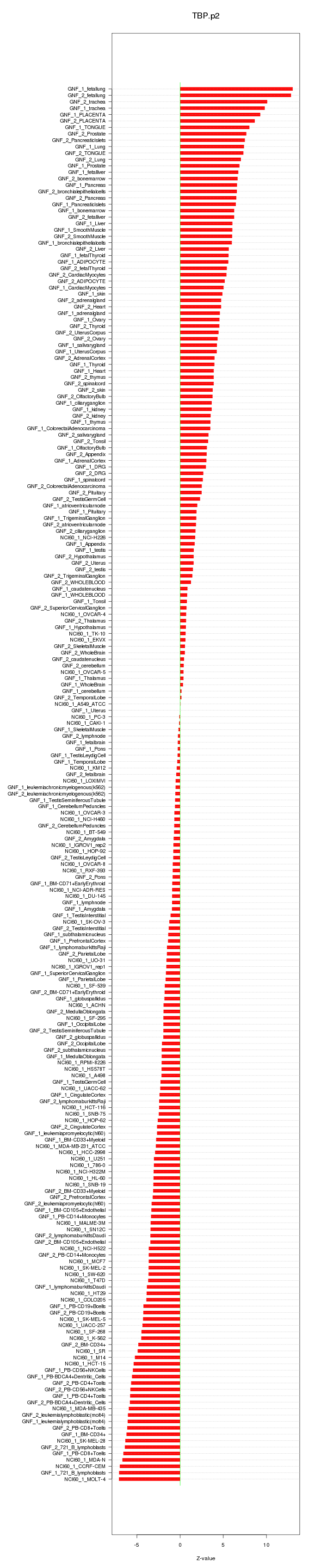
|
2.44
|
4.99e-26
|
GO:0042221
|
response to chemical stimulus
|
|
2.24
|
2.11e-21
|
GO:0006950
|
response to stress
|
|
3.24
|
4.86e-17
|
GO:0006952
|
defense response
|
|
2.58
|
5.88e-15
|
GO:0010033
|
response to organic substance
|
|
4.98
|
7.92e-15
|
GO:0006954
|
inflammatory response
|
|
1.57
|
2.32e-14
|
GO:0032501
|
multicellular organismal process
|
|
2.20
|
2.36e-14
|
GO:0048513
|
organ development
|
|
4.86
|
1.22e-13
|
GO:0010035
|
response to inorganic substance
|
|
2.86
|
1.59e-13
|
GO:0009611
|
response to wounding
|
|
1.47
|
3.27e-13
|
GO:0050896
|
response to stimulus
|
|
1.69
|
1.28e-12
|
GO:0032502
|
developmental process
|
|
2.83
|
2.18e-12
|
GO:0009888
|
tissue development
|
|
2.01
|
3.87e-12
|
GO:0065008
|
regulation of biological quality
|
|
2.71
|
4.15e-12
|
GO:0070887
|
cellular response to chemical stimulus
|
|
8.07
|
4.39e-12
|
GO:0006334
|
nucleosome assembly
|
|
1.77
|
6.02e-12
|
GO:0048856
|
anatomical structure development
|
|
7.66
|
1.53e-11
|
GO:0031497
|
chromatin assembly
|
|
7.28
|
4.92e-11
|
GO:0034728
|
nucleosome organization
|
|
7.01
|
1.19e-10
|
GO:0065004
|
protein-DNA complex assembly
|
|
1.78
|
1.64e-10
|
GO:0048731
|
system development
|
|
3.29
|
3.52e-10
|
GO:0009607
|
response to biotic stimulus
|
|
4.46
|
4.10e-10
|
GO:0009617
|
response to bacterium
|
|
6.63
|
4.15e-10
|
GO:0071824
|
protein-DNA complex subunit organization
|
|
1.66
|
5.96e-10
|
GO:0007275
|
multicellular organismal development
|
|
6.10
|
8.51e-10
|
GO:0006323
|
DNA packaging
|
|
2.56
|
1.74e-09
|
GO:0051704
|
multi-organism process
|
|
3.49
|
1.95e-09
|
GO:0051707
|
response to other organism
|
|
2.24
|
2.17e-09
|
GO:0051239
|
regulation of multicellular organismal process
|
|
7.26
|
2.63e-09
|
GO:0000302
|
response to reactive oxygen species
|
|
2.92
|
5.55e-09
|
GO:0009725
|
response to hormone stimulus
|
|
3.32
|
5.77e-09
|
GO:0051094
|
positive regulation of developmental process
|
|
2.48
|
1.01e-08
|
GO:0050793
|
regulation of developmental process
|
|
10.76
|
1.88e-08
|
GO:0034614
|
cellular response to reactive oxygen species
|
|
5.50
|
2.50e-08
|
GO:0006333
|
chromatin assembly or disassembly
|
|
5.24
|
2.65e-08
|
GO:0071103
|
DNA conformation change
|
|
2.83
|
2.86e-08
|
GO:0048878
|
chemical homeostasis
|
|
8.61
|
2.88e-08
|
GO:0034599
|
cellular response to oxidative stress
|
|
2.71
|
5.09e-08
|
GO:0009719
|
response to endogenous stimulus
|
|
4.64
|
6.26e-08
|
GO:0006979
|
response to oxidative stress
|
|
2.44
|
6.77e-08
|
GO:0042592
|
homeostatic process
|
|
7.95
|
1.09e-07
|
GO:0042542
|
response to hydrogen peroxide
|
|
2.56
|
1.15e-07
|
GO:0006955
|
immune response
|
|
4.42
|
1.89e-07
|
GO:0008544
|
epidermis development
|
|
2.61
|
2.07e-07
|
GO:0002682
|
regulation of immune system process
|
|
2.31
|
2.72e-07
|
GO:0042127
|
regulation of cell proliferation
|
|
3.80
|
3.10e-07
|
GO:0048545
|
response to steroid hormone stimulus
|
|
12.69
|
4.19e-07
|
GO:0070301
|
cellular response to hydrogen peroxide
|
|
1.66
|
4.99e-07
|
GO:0048518
|
positive regulation of biological process
|
|
2.09
|
7.79e-07
|
GO:0002376
|
immune system process
|
|
8.37
|
8.32e-07
|
GO:0002526
|
acute inflammatory response
|
|
2.25
|
8.95e-07
|
GO:0009605
|
response to external stimulus
|
|
5.59
|
3.34e-06
|
GO:0008217
|
regulation of blood pressure
|
|
2.62
|
3.77e-06
|
GO:0009887
|
organ morphogenesis
|
|
3.85
|
3.95e-06
|
GO:0032101
|
regulation of response to external stimulus
|
|
2.38
|
7.34e-06
|
GO:2000026
|
regulation of multicellular organismal development
|
|
2.25
|
1.28e-05
|
GO:0040011
|
locomotion
|
|
2.26
|
1.45e-05
|
GO:0043933
|
macromolecular complex subunit organization
|
|
3.07
|
1.71e-05
|
GO:0034622
|
cellular macromolecular complex assembly
|
|
4.55
|
1.72e-05
|
GO:0002237
|
response to molecule of bacterial origin
|
|
2.74
|
2.42e-05
|
GO:0009628
|
response to abiotic stimulus
|
|
12.11
|
3.15e-05
|
GO:0042743
|
hydrogen peroxide metabolic process
|
|
2.57
|
3.58e-05
|
GO:0006928
|
cellular component movement
|
|
4.14
|
4.10e-05
|
GO:0010038
|
response to metal ion
|
|
1.24
|
4.70e-05
|
GO:0065007
|
biological regulation
|
|
3.07
|
5.06e-05
|
GO:0051240
|
positive regulation of multicellular organismal process
|
|
2.66
|
5.25e-05
|
GO:0034621
|
cellular macromolecular complex subunit organization
|
|
1.61
|
6.39e-05
|
GO:0048522
|
positive regulation of cellular process
|
|
3.32
|
8.96e-05
|
GO:0006875
|
cellular metal ion homeostasis
|
|
3.04
|
1.04e-04
|
GO:0045597
|
positive regulation of cell differentiation
|
|
1.41
|
1.45e-04
|
GO:0051716
|
cellular response to stimulus
|
|
1.81
|
1.54e-04
|
GO:0009653
|
anatomical structure morphogenesis
|
|
2.62
|
1.81e-04
|
GO:0072358
|
cardiovascular system development
|
|
2.62
|
1.81e-04
|
GO:0072359
|
circulatory system development
|
|
3.21
|
1.82e-04
|
GO:0055065
|
metal ion homeostasis
|
|
4.46
|
3.00e-04
|
GO:0051789
|
response to protein stimulus
|
|
5.05
|
3.15e-04
|
GO:0009266
|
response to temperature stimulus
|
|
2.24
|
3.83e-04
|
GO:0065003
|
macromolecular complex assembly
|
|
8.07
|
3.99e-04
|
GO:0072593
|
reactive oxygen species metabolic process
|
|
3.61
|
4.43e-04
|
GO:0003013
|
circulatory system process
|
|
3.61
|
4.43e-04
|
GO:0008015
|
blood circulation
|
|
4.07
|
5.48e-04
|
GO:0042445
|
hormone metabolic process
|
|
4.26
|
6.04e-04
|
GO:0032496
|
response to lipopolysaccharide
|
|
3.01
|
6.30e-04
|
GO:0030003
|
cellular cation homeostasis
|
|
7.69
|
6.57e-04
|
GO:0031424
|
keratinization
|
|
7.69
|
6.57e-04
|
GO:0045639
|
positive regulation of myeloid cell differentiation
|
|
12.56
|
9.92e-04
|
GO:0042744
|
hydrogen peroxide catabolic process
|
|
2.21
|
1.04e-03
|
GO:0045595
|
regulation of cell differentiation
|
|
2.85
|
1.11e-03
|
GO:0055080
|
cation homeostasis
|
|
4.57
|
1.19e-03
|
GO:0050727
|
regulation of inflammatory response
|
|
3.25
|
1.45e-03
|
GO:0008202
|
steroid metabolic process
|
|
2.53
|
1.49e-03
|
GO:0055082
|
cellular chemical homeostasis
|
|
3.03
|
1.61e-03
|
GO:0001817
|
regulation of cytokine production
|
|
2.35
|
1.66e-03
|
GO:0019725
|
cellular homeostasis
|
|
3.46
|
1.73e-03
|
GO:0010817
|
regulation of hormone levels
|
|
5.12
|
1.79e-03
|
GO:0045637
|
regulation of myeloid cell differentiation
|
|
1.54
|
1.83e-03
|
GO:0048519
|
negative regulation of biological process
|
|
3.57
|
1.98e-03
|
GO:0003012
|
muscle system process
|
|
7.86
|
2.12e-03
|
GO:0030049
|
muscle filament sliding
|
|
7.86
|
2.12e-03
|
GO:0033275
|
actin-myosin filament sliding
|
|
3.68
|
2.44e-03
|
GO:0006936
|
muscle contraction
|
|
20.18
|
2.47e-03
|
GO:0042738
|
exogenous drug catabolic process
|
|
6.73
|
2.49e-03
|
GO:0045766
|
positive regulation of angiogenesis
|
|
1.80
|
2.56e-03
|
GO:0009893
|
positive regulation of metabolic process
|
|
3.51
|
2.58e-03
|
GO:0030855
|
epithelial cell differentiation
|
|
7.65
|
2.71e-03
|
GO:0070252
|
actin-mediated cell contraction
|
|
2.55
|
2.77e-03
|
GO:0016477
|
cell migration
|
|
2.85
|
2.86e-03
|
GO:0034097
|
response to cytokine stimulus
|
|
13.84
|
3.44e-03
|
GO:0071347
|
cellular response to interleukin-1
|
|
2.35
|
3.45e-03
|
GO:0006935
|
chemotaxis
|
|
2.35
|
3.45e-03
|
GO:0042330
|
taxis
|
|
2.89
|
3.66e-03
|
GO:0040012
|
regulation of locomotion
|
|
5.24
|
3.68e-03
|
GO:0009913
|
epidermal cell differentiation
|
|
3.06
|
3.84e-03
|
GO:0051241
|
negative regulation of multicellular organismal process
|
|
1.87
|
3.87e-03
|
GO:0022607
|
cellular component assembly
|
|
2.61
|
4.08e-03
|
GO:0060429
|
epithelium development
|
|
2.46
|
4.14e-03
|
GO:0002684
|
positive regulation of immune system process
|
|
3.71
|
4.29e-03
|
GO:0050900
|
leukocyte migration
|
|
2.33
|
4.38e-03
|
GO:0008284
|
positive regulation of cell proliferation
|
|
6.33
|
4.48e-03
|
GO:0030048
|
actin filament-based movement
|
|
1.57
|
4.54e-03
|
GO:0030154
|
cell differentiation
|
|
4.72
|
4.66e-03
|
GO:0001818
|
negative regulation of cytokine production
|
|
2.85
|
4.75e-03
|
GO:0042493
|
response to drug
|
|
2.69
|
5.26e-03
|
GO:0043434
|
response to peptide hormone stimulus
|
|
2.06
|
5.35e-03
|
GO:0071844
|
cellular component assembly at cellular level
|
|
5.55
|
5.49e-03
|
GO:0030216
|
keratinocyte differentiation
|
|
2.74
|
6.07e-03
|
GO:0009991
|
response to extracellular stimulus
|
|
2.45
|
6.11e-03
|
GO:0006873
|
cellular ion homeostasis
|
|
2.40
|
6.83e-03
|
GO:0048870
|
cell motility
|
|
2.40
|
6.83e-03
|
GO:0051674
|
localization of cell
|
|
2.00
|
6.87e-03
|
GO:0048584
|
positive regulation of response to stimulus
|
|
2.94
|
7.69e-03
|
GO:0030334
|
regulation of cell migration
|
|
1.55
|
7.71e-03
|
GO:0048869
|
cellular developmental process
|
|
5.98
|
7.75e-03
|
GO:0051591
|
response to cAMP
|
|
2.68
|
8.69e-03
|
GO:0031347
|
regulation of defense response
|
|
4.47
|
8.71e-03
|
GO:0007586
|
digestion
|
|
7.83
|
8.78e-03
|
GO:0070555
|
response to interleukin-1
|
|
3.91
|
8.93e-03
|
GO:0031960
|
response to corticosteroid stimulus
|
|
1.11
|
8.94e-03
|
GO:0009987
|
cellular process
|
|
2.90
|
9.40e-03
|
GO:2000145
|
regulation of cell motility
|
|
4.42
|
9.83e-03
|
GO:0045765
|
regulation of angiogenesis
|
|
2.17
|
1.04e-02
|
GO:0042060
|
wound healing
|
|
16.15
|
1.06e-02
|
GO:0001542
|
ovulation from ovarian follicle
|
|
16.15
|
1.06e-02
|
GO:0042737
|
drug catabolic process
|
|
16.15
|
1.06e-02
|
GO:0045672
|
positive regulation of osteoclast differentiation
|
|
4.37
|
1.11e-02
|
GO:0032103
|
positive regulation of response to external stimulus
|
|
2.34
|
1.11e-02
|
GO:0050801
|
ion homeostasis
|
|
9.04
|
1.25e-02
|
GO:0002763
|
positive regulation of myeloid leukocyte differentiation
|
|
1.75
|
1.53e-02
|
GO:0031325
|
positive regulation of cellular metabolic process
|
|
2.72
|
1.66e-02
|
GO:0031667
|
response to nutrient levels
|
|
3.93
|
1.72e-02
|
GO:0051384
|
response to glucocorticoid stimulus
|
|
3.90
|
1.91e-02
|
GO:0042742
|
defense response to bacterium
|
|
5.38
|
2.08e-02
|
GO:0033500
|
carbohydrate homeostasis
|
|
5.38
|
2.08e-02
|
GO:0042593
|
glucose homeostasis
|
|
8.37
|
2.19e-02
|
GO:0031032
|
actomyosin structure organization
|
|
6.98
|
2.20e-02
|
GO:0006953
|
acute-phase response
|
|
3.62
|
2.33e-02
|
GO:0009410
|
response to xenobiotic stimulus
|
|
5.93
|
2.55e-02
|
GO:0055002
|
striated muscle cell development
|
|
1.73
|
2.61e-02
|
GO:0044085
|
cellular component biogenesis
|
|
4.74
|
2.69e-02
|
GO:0002576
|
platelet degranulation
|
|
1.74
|
2.70e-02
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
|
8.07
|
2.85e-02
|
GO:0032963
|
collagen metabolic process
|
|
3.33
|
3.37e-02
|
GO:0070482
|
response to oxygen levels
|
|
9.69
|
3.81e-02
|
GO:0045103
|
intermediate filament-based process
|
|
2.73
|
4.02e-02
|
GO:0001568
|
blood vessel development
|
|
2.64
|
4.14e-02
|
GO:0051270
|
regulation of cellular component movement
|
|
4.97
|
4.32e-02
|
GO:0019748
|
secondary metabolic process
|
|
7.54
|
4.65e-02
|
GO:0044259
|
multicellular organismal macromolecule metabolic process
|
|
4.12
|
4.66e-02
|
GO:0030198
|
extracellular matrix organization
|
|
6.30
|
4.90e-02
|
GO:0006721
|
terpenoid metabolic process
|
|
3.59
|
4.94e-02
|
GO:0043627
|
response to estrogen stimulus
|