|
chr20_+_10199455
|
11.108
|
NM_003081
NM_130811
|
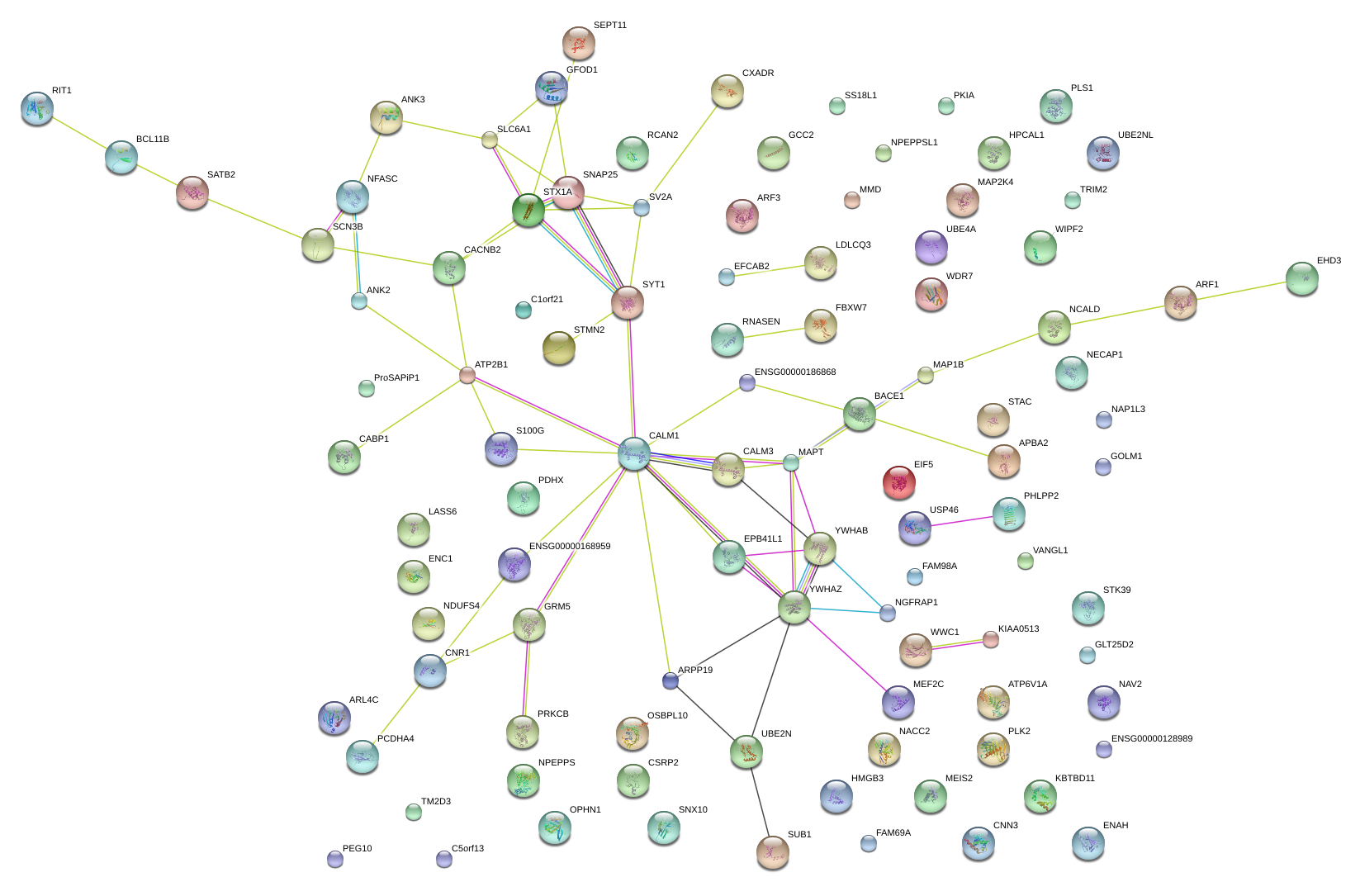
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr8_+_80523048
|
10.534
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr5_-_57755787
|
10.325
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr2_-_200335988
|
9.155
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr5_-_73937248
|
8.777
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr2_-_200329810
|
8.621
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr7_+_94285676
|
8.423
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chrX_+_102631267
|
7.906
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr2_-_200322699
|
7.899
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chrX_+_102632108
|
7.883
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr8_-_103137134
|
7.822
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr7_+_94285620
|
7.741
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr16_+_23847271
|
7.576
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr12_+_79439432
|
7.510
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chrX_-_92928566
|
7.465
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr12_+_79257772
|
7.385
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr6_-_46293628
|
7.335
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr12_+_79258448
|
7.305
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr5_+_71403040
|
7.164
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr8_-_103136486
|
6.911
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr1_-_149889362
|
6.861
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr1_-_95392501
|
6.526
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr4_-_53522756
|
6.140
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr8_-_102803438
|
6.117
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr20_+_34700257
|
6.038
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr12_-_49351251
|
6.023
|
NM_001659
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr7_-_73133917
|
5.918
|
NM_001165903
NM_004603
|
STX1A
|
syntaxin 1A (brain)
|
|
chr20_+_34742656
|
5.761
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr2_+_109065576
|
5.705
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr11_+_34937676
|
5.554
|
NM_001135024
NM_001166158
NM_003477
|
PDHX
|
pyruvate dehydrogenase complex, component X
|
|
chr17_-_53499055
|
5.518
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr11_-_117166247
|
5.507
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr4_-_53525316
|
5.459
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr12_+_121088370
|
5.433
|
NM_004276
NM_031205
|
CABP1
|
calcium binding protein 1
|
|
chr8_+_79428335
|
5.382
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr19_+_47104490
|
5.329
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr1_+_204913353
|
5.323
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr9_-_138987096
|
5.307
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr11_-_123525300
|
5.299
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr15_+_29213839
|
5.236
|
NM_001130414
NM_005503
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2
|
|
chr12_+_121078421
|
5.176
|
NM_001033677
|
CABP1
|
calcium binding protein 1
|
|
chr15_-_102192337
|
5.132
|
NM_025141
NM_078474
|
TM2D3
|
TM2 domain containing 3
|
|
chr17_+_45608443
|
4.875
|
NM_006310
|
NPEPPS
|
aminopeptidase puromycin sensitive
|
|
chr12_-_90049818
|
4.849
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chrX_+_150151757
|
4.846
|
NM_005342
|
HMGB3
|
high mobility group box 3
|
|
chr4_-_153303663
|
4.822
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr2_+_31456879
|
4.770
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr15_-_52861177
|
4.721
|
NM_006628
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr8_+_1922030
|
4.588
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr2_+_169312793
|
4.585
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chr11_+_19372270
|
4.455
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chrX_-_67653169
|
4.454
|
NM_002547
|
OPHN1
|
oligophrenin 1
|
|
chr3_+_11034419
|
4.450
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr11_+_118230292
|
4.445
|
NM_001204077
NM_004788
|
UBE4A
|
ubiquitination factor E4A
|
|
chr15_-_37393364
|
4.388
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr20_-_3149070
|
4.337
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr4_-_153456152
|
4.326
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr10_-_62493282
|
4.301
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr5_-_111093792
|
4.288
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_74632992
|
4.256
|
NM_000859
NM_001130996
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase
|
|
chr4_+_154125589
|
4.252
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr3_+_113465860
|
4.242
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr1_-_155881163
|
4.185
|
NM_006912
|
RIT1
|
Ras-like without CAAX 1
|
|
chr15_-_37390372
|
4.180
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr4_+_77870864
|
4.102
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr18_+_54318561
|
4.042
|
NM_015285
NM_052834
|
WDR7
|
WD repeat domain 7
|
|
chr4_+_154074269
|
4.019
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_-_169103886
|
4.014
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr16_+_85061303
|
3.985
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr17_+_43971655
|
3.967
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr21_+_18885104
|
3.967
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr9_-_88714486
|
3.911
|
NM_016548
|
GOLM1
|
golgi membrane protein 1
|
|
chr12_+_8234796
|
3.899
|
NM_015509
|
NECAP1
|
NECAP endocytosis associated 1
|
|
chr12_-_77272767
|
3.882
|
NM_001321
|
CSRP2
|
cysteine and glycine-rich protein 2
|
|
chr17_+_11924116
|
3.881
|
NM_003010
|
MAP2K4
|
mitogen-activated protein kinase kinase 4
|
|
chr5_-_111093251
|
3.876
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_184356128
|
3.760
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr20_+_43514340
|
3.734
|
NM_003404
NM_139323
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr7_+_26403944
|
3.676
|
NM_001199838
|
SNX10
|
sorting nexin 10
|
|
chr11_+_19734821
|
3.674
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr11_-_117186918
|
3.660
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr12_-_93835986
|
3.626
|
NM_003348
|
UBE2N
|
ubiquitin-conjugating enzyme E2N
|
|
chr2_-_235405692
|
3.565
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr1_-_225840660
|
3.562
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr9_-_88715024
|
3.527
|
NM_177937
|
GOLM1
|
golgi membrane protein 1
|
|
chr4_+_113739203
|
3.518
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr5_-_88179278
|
3.485
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr5_-_111093030
|
3.478
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_32023237
|
3.478
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr1_+_116185249
|
3.474
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr10_+_18689512
|
3.444
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr1_-_184006828
|
3.430
|
NM_015101
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr20_+_60718473
|
3.417
|
NM_198935
|
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr1_-_93426998
|
3.402
|
NM_001006605
NM_001252269
NM_001252270
NM_001252273
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr15_-_37391596
|
3.395
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr6_-_88875766
|
3.393
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr14_+_103801160
|
3.382
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr5_+_52856460
|
3.327
|
NM_002495
|
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase)
|
|
chr6_-_13408368
|
3.318
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr5_+_32585604
|
3.278
|
NM_006713
|
SUB1
|
SUB1 homolog (S. cerevisiae)
|
|
chr5_-_111312522
|
3.250
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_-_33824294
|
3.244
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr7_+_26331493
|
3.224
|
NM_001199835
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr1_+_116184573
|
3.219
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr16_-_71748697
|
3.215
|
NM_015020
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2
|
|
chr5_+_167718485
|
3.177
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr7_+_26400503
|
3.159
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr11_-_88796815
|
3.144
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr3_+_36421944
|
3.123
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr3_+_142375738
|
3.120
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr5_-_31532164
|
3.113
|
NM_001100412
NM_013235
|
DROSHA
|
drosha, ribonuclease type III
|
|
chr17_+_38375554
|
3.111
|
NM_133264
|
WIPF2
|
WAS/WASL interacting protein family, member 2
|
|
chr11_+_16760147
|
3.108
|
NM_014267
|
C11orf58
|
chromosome 11 open reading frame 58
|
|
chr10_-_62332666
|
3.079
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_-_86043932
|
3.075
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr15_-_37392358
|
3.072
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr3_-_134093235
|
3.054
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr7_-_108096693
|
2.995
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr15_-_42749710
|
2.993
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr21_-_43346780
|
2.987
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr19_-_49576120
|
2.984
|
NM_031886
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr4_+_113970784
|
2.972
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr6_-_143266283
|
2.938
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chrX_-_6146655
|
2.922
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr1_+_204797774
|
2.909
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr5_-_88199868
|
2.909
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_-_139163222
|
2.905
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr21_-_43373938
|
2.898
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr5_+_112312392
|
2.883
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr10_-_62149487
|
2.863
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_-_85930733
|
2.851
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr6_+_111195986
|
2.844
|
NM_001634
NM_001033059
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr11_-_45687135
|
2.842
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr4_-_48782281
|
2.836
|
NM_015030
|
FRYL
|
FRY-like
|
|
chr22_+_46067677
|
2.804
|
NM_001167621
NM_013236
|
ATXN10
|
ataxin 10
|
|
chr5_-_43313581
|
2.799
|
NM_001098272
NM_002130
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr17_-_46178559
|
2.743
|
NM_006807
|
CBX1
|
chromobox homolog 1
|
|
chr10_+_22610138
|
2.742
|
NM_005180
|
BMI1
|
BMI1 polycomb ring finger oncogene
|
|
chr11_-_128894008
|
2.710
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr16_+_6069094
|
2.693
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_-_95007139
|
2.687
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr17_-_8534008
|
2.675
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr3_+_142315205
|
2.660
|
NM_001145319
|
PLS1
|
plastin 1
|
|
chrX_-_6145887
|
2.654
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr5_-_111091947
|
2.650
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_+_142342265
|
2.630
|
NM_002670
|
PLS1
|
plastin 1
|
|
chr17_+_28707495
|
2.600
|
NM_001199775
|
CPD
|
carboxypeptidase D
|
|
chr18_-_812268
|
2.590
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr10_-_21463019
|
2.562
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr10_+_111765725
|
2.547
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr17_+_42385780
|
2.542
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr17_+_15848230
|
2.529
|
NM_000676
|
ADORA2B
|
adenosine A2b receptor
|
|
chr20_-_48732479
|
2.505
|
NM_022442
NM_021988
NM_199144
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr10_+_111767685
|
2.491
|
NM_001121
NM_016824
|
ADD3
|
adducin 3 (gamma)
|
|
chr20_-_33999780
|
2.473
|
NM_001184977
NM_018244
NM_199487
|
UQCC
|
ubiquinol-cytochrome c reductase complex chaperone
|
|
chr2_+_7057471
|
2.473
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr12_-_124457099
|
2.447
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr17_+_72983723
|
2.446
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr6_-_13486382
|
2.438
|
NM_001242628
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr17_+_30813928
|
2.433
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr16_-_4292052
|
2.432
|
NM_001098814
|
SRL
|
sarcalumenin
|
|
chr12_-_54653369
|
2.429
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr1_+_193028523
|
2.424
|
NM_001173524
|
TROVE2
|
TROVE domain family, member 2
|
|
chr14_+_92790151
|
2.417
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr5_-_111092918
|
2.394
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_+_142622797
|
2.393
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr15_-_73925658
|
2.348
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr10_+_18629613
|
2.298
|
NM_201590
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr11_+_73882316
|
2.289
|
NM_016147
|
PPME1
|
protein phosphatase methylesterase 1
|
|
chr14_-_103523619
|
2.286
|
NM_006035
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like)
|
|
chr4_-_71705613
|
2.284
|
NM_002092
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr3_+_193310921
|
2.283
|
NM_015560
NM_130831
NM_130832
NM_130833
NM_130834
NM_130835
NM_130836
NM_130837
|
OPA1
|
optic atrophy 1 (autosomal dominant)
|
|
chr1_-_222885653
|
2.262
|
NM_022831
|
AIDA
|
axin interactor, dorsalization associated
|
|
chr4_+_174089889
|
2.256
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr2_+_103089761
|
2.223
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr17_-_46178761
|
2.219
|
NM_001127228
|
CBX1
|
chromobox homolog 1
|
|
chr1_-_68299047
|
2.207
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr4_-_122744978
|
2.195
|
NM_001237
|
CCNA2
|
cyclin A2
|
|
chrX_+_154444700
|
2.195
|
NM_003372
|
VBP1
|
von Hippel-Lindau binding protein 1
|
|
chr1_-_78148326
|
2.195
|
NM_015534
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr11_+_34642587
|
2.185
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr4_-_71705188
|
2.170
|
NM_001098477
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr3_-_56502364
|
2.143
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr6_-_119470336
|
2.140
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr14_+_73603140
|
2.140
|
NM_000021
NM_007318
|
PSEN1
|
presenilin 1
|
|
chr1_+_65613771
|
2.136
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr17_-_40075238
|
2.134
|
NM_001096
NM_198830
|
ACLY
|
ATP citrate lyase
|
|
chr20_-_48729664
|
2.130
|
NM_001032288
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr16_+_6823809
|
2.128
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr22_-_44258210
|
2.112
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr11_-_129062092
|
2.109
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr10_+_94050858
|
2.105
|
NM_017824
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5
|
|
chr3_+_54156691
|
2.095
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr10_+_18429605
|
2.084
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr4_-_153273683
|
2.057
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr9_+_117373622
|
2.055
|
NM_153045
|
C9orf91
|
chromosome 9 open reading frame 91
|
|
chr16_-_70719854
|
2.039
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr10_-_91405320
|
2.036
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr11_+_58939539
|
2.013
|
NM_015177
|
DTX4
|
deltex homolog 4 (Drosophila)
|
|
chr1_+_151512760
|
1.995
|
NM_001126337
NM_020127
|
TUFT1
|
tuftelin 1
|