|
chr2_+_203499819
|
32.387
|
NM_173511
|
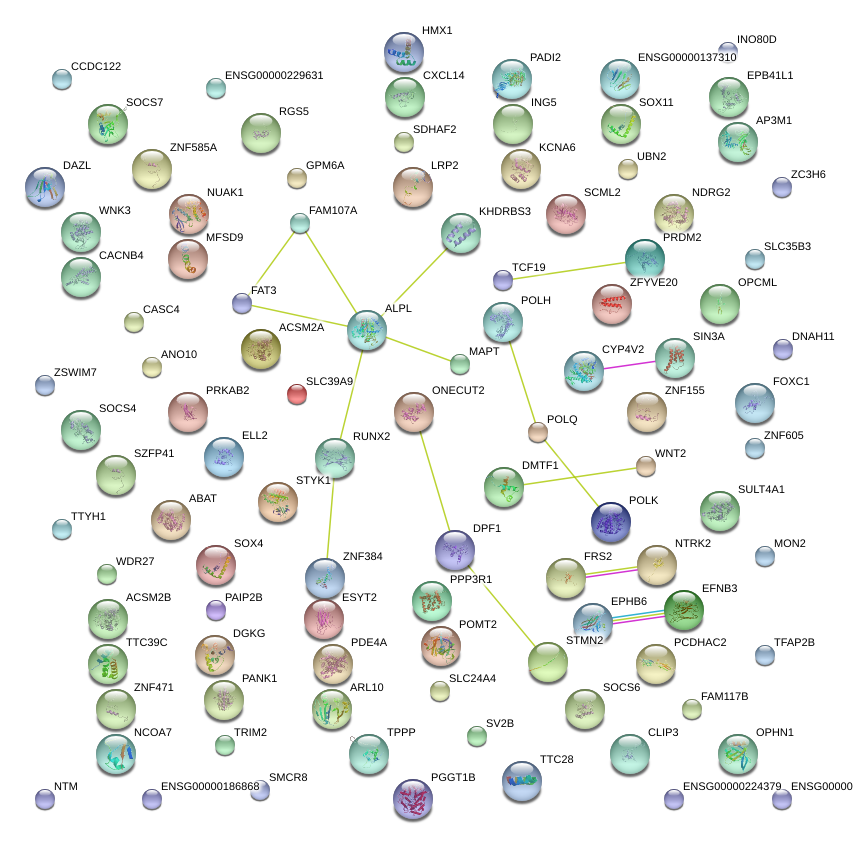
FAM117B
|
family with sequence similarity 117, member B
|
|
chr18_+_21594349
|
28.681
|
NM_001135993
NM_001243425
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr10_-_75910525
|
26.076
|
NM_012095
NM_207012
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit
|
|
chr5_-_693413
|
25.684
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr7_+_142552547
|
21.840
|
NM_004445
|
EPHB6
|
EPH receptor B6
|
|
chr18_+_21572736
|
20.470
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr18_-_13726582
|
19.167
|
NM_001098801
NM_152352
|
FAM210A
|
family with sequence similarity 210, member A
|
|
chr2_-_206950895
|
17.957
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr6_+_1610680
|
17.160
|
NM_001453
|
FOXC1
|
forkhead box C1
|
|
chr7_+_138916230
|
15.370
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr3_-_16646244
|
15.060
|
NM_001190811
|
DAZL
|
deleted in azoospermia-like
|
|
chrX_-_18372777
|
14.863
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr9_+_87283453
|
14.628
|
NM_001018064
NM_006180
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr11_+_92085261
|
14.471
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr19_+_54926604
|
14.432
|
NM_001005367
NM_001201461
NM_020659
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr17_+_7608413
|
14.411
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr7_-_158622202
|
14.240
|
NM_020728
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr19_-_38720312
|
14.190
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr3_-_16646927
|
13.762
|
NM_001351
|
DAZL
|
deleted in azoospermia-like
|
|
chr6_+_126111782
|
13.461
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr2_-_71454200
|
13.349
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr17_+_36507708
|
13.316
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr17_-_15902907
|
13.176
|
NM_001042697
NM_001042698
|
ZSWIM7
|
zinc finger, SWIM-type containing 7
|
|
chr6_+_126240369
|
13.130
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr17_+_18218569
|
12.792
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr12_+_69864065
|
12.510
|
NM_001042555
NM_006654
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr6_-_8435624
|
12.444
|
NM_001142540
NM_001142541
NM_015948
|
SLC35B3
|
solute carrier family 35, member B3
|
|
chr6_+_126102278
|
12.432
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr12_-_6798619
|
12.382
|
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr19_-_37701385
|
12.216
|
NM_152279
|
ZNF585B
|
zinc finger protein 585B
|
|
chr5_-_95297597
|
12.028
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr3_-_58563490
|
11.961
|
NM_001076778
NM_007177
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr11_+_17373138
|
11.674
|
NM_001202439
|
B7H6
|
B7 homolog 6
|
|
chr12_-_6798528
|
11.588
|
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr13_-_44453825
|
11.275
|
NM_144974
|
CCDC122
|
coiled-coil domain containing 122
|
|
chr19_-_37019169
|
11.108
|
NM_001012756
NM_001166036
NM_001166037
NM_001166038
|
ZNF260
|
zinc finger protein 260
|
|
chr20_+_34742656
|
11.054
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr4_+_187112635
|
10.968
|
NM_207352
|
CYP4V2
|
cytochrome P450, family 4, subfamily V, polypeptide 2
|
|
chr17_+_43971655
|
10.939
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr16_+_8768403
|
10.839
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr2_+_113033146
|
10.335
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr15_+_44580870
|
10.164
|
NM_138423
NM_177974
|
CASC4
|
cancer susceptibility candidate 4
|
|
chr14_+_69865380
|
10.159
|
NM_001252148
NM_001252150
NM_018375
|
SLC39A9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr18_+_55102777
|
10.152
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr1_-_146644045
|
10.099
|
NM_005399
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit
|
|
chr1_-_163172624
|
10.086
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr12_+_62860531
|
10.041
|
NM_015026
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr14_+_92790151
|
9.966
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr7_+_86781869
|
9.955
|
NM_021145
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr4_-_176734181
|
9.871
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr2_-_103353300
|
9.795
|
NM_032718
|
MFSD9
|
major facilitator superfamily domain containing 9
|
|
chr1_+_21835841
|
9.731
|
NM_000478
NM_001127501
NM_001177520
|
ALPL
|
alkaline phosphatase, liver/bone/kidney
|
|
chr15_+_91643429
|
9.612
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr6_+_45389818
|
9.559
|
NM_004348
|
RUNX2
|
runt-related transcription factor 2
|
|
chrX_-_67653169
|
9.510
|
NM_002547
|
OPHN1
|
oligophrenin 1
|
|
chr4_+_175204827
|
9.456
|
NM_001040157
|
CEP44
|
centrosomal protein 44kDa
|
|
chr1_-_163172863
|
9.451
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr19_+_57019196
|
9.413
|
NM_020813
|
ZNF471
|
zinc finger protein 471
|
|
chr1_+_14026685
|
9.336
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr2_-_170219012
|
9.326
|
NM_004525
|
LRP2
|
low density lipoprotein receptor-related protein 2
|
|
chr16_+_20462858
|
9.303
|
NM_001010845
|
ACSM2A
|
acyl-CoA synthetase medium-chain family member 2A
|
|
chr6_-_170102098
|
9.294
|
NM_001202550
NM_182552
|
WDR27
|
WD repeat domain 27
|
|
chr3_-_15140652
|
9.236
|
NM_022340
|
ZFYVE20
|
zinc finger, FYVE domain containing 20
|
|
chr2_-_152830448
|
9.228
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr12_-_133532847
|
9.195
|
NM_183238
NM_001164715
|
ZNF605
|
zinc finger protein 605
|
|
chr19_-_36523540
|
9.132
|
NM_001199570
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr5_-_134914968
|
9.088
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr12_+_4918341
|
9.067
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr7_-_116963328
|
9.026
|
NM_003391
|
WNT2
|
wingless-type MMTV integration site family member 2
|
|
chr11_-_133402227
|
9.009
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr7_+_86781644
|
8.899
|
NM_001142326
NM_001142327
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr12_-_10826890
|
8.861
|
NM_018423
|
STYK1
|
serine/threonine/tyrosine kinase 1
|
|
chr14_+_92789536
|
8.835
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr4_-_8873453
|
8.813
|
NM_018942
|
HMX1
|
H6 family homeobox 1
|
|
chr8_+_80523048
|
8.742
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr11_+_61197574
|
8.661
|
NM_017841
|
SDHAF2
|
succinate dehydrogenase complex assembly factor 2
|
|
chr19_+_44488331
|
8.636
|
NM_003445
NM_198089
|
ZNF155
|
zinc finger protein 155
|
|
chr2_+_242641310
|
8.635
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr10_-_91403630
|
8.634
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr3_-_186079776
|
8.618
|
NM_001080744
NM_001080745
NM_001346
|
DGKG
|
diacylglycerol kinase, gamma 90kDa
|
|
chr4_+_175205044
|
8.607
|
NM_001145314
|
CEP44
|
centrosomal protein 44kDa
|
|
chr15_-_75743776
|
8.597
|
NM_015477
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr22_-_44258210
|
8.567
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr6_+_31126114
|
8.543
|
NM_001077511
NM_007109
|
TCF19
|
transcription factor 19
|
|
chr5_+_175792444
|
8.540
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr1_-_17445865
|
8.467
|
NM_007365
|
PADI2
|
peptidyl arginine deiminase, type II
|
|
chr8_+_136469557
|
8.427
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr4_+_154074269
|
8.369
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr22_-_29075708
|
8.339
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr19_-_36523770
|
8.326
|
NM_015526
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr2_-_68479481
|
8.292
|
NM_000945
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr5_-_114598547
|
8.188
|
NM_005023
|
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit
|
|
chr11_-_132812870
|
8.187
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr15_+_91643181
|
8.090
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr19_+_10541334
|
8.073
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr2_-_152955208
|
7.961
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr2_+_5832778
|
7.901
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr6_+_50786438
|
7.899
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chrX_-_54384436
|
7.898
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr14_-_21493910
|
7.883
|
NM_016250
NM_201535
NM_201536
NM_201537
|
NDRG2
|
NDRG family member 2
|
|
chr5_+_140345746
|
7.833
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr14_-_77787023
|
7.821
|
NM_013382
|
POMT2
|
protein-O-mannosyltransferase 2
|
|
chr12_-_106533810
|
7.815
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr16_+_8806825
|
7.788
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr6_+_43543877
|
7.755
|
NM_006502
|
POLH
|
polymerase (DNA directed), eta
|
|
chr6_+_39016556
|
7.746
|
NM_002062
|
GLP1R
|
glucagon-like peptide 1 receptor
|
|
chr19_+_57752051
|
7.738
|
NM_001023563
NM_001145078
|
ZNF805
|
zinc finger protein 805
|
|
chr4_-_176923572
|
7.734
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr1_+_183605207
|
7.671
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr6_+_126070724
|
7.543
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr15_-_50978983
|
7.489
|
NM_017672
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr20_+_34700257
|
7.479
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr3_-_122134881
|
7.419
|
NM_019069
|
WDR5B
|
WD repeat domain 5B
|
|
chr15_-_75748123
|
7.416
|
NM_001145357
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr8_-_17271015
|
7.383
|
NM_004686
|
MTMR7
|
myotubularin related protein 7
|
|
chr6_+_147829784
|
7.381
|
NM_001030060
|
SAMD5
|
sterile alpha motif domain containing 5
|
|
chr9_-_35665204
|
7.376
|
NM_032818
|
C9orf100
|
chromosome 9 open reading frame 100
|
|
chr12_+_133757994
|
7.349
|
NM_001165883
NM_001165884
NM_001165885
NM_001165886
NM_001165887
NM_003415
NM_152943
NM_001165881
NM_001165882
|
ZNF268
|
zinc finger protein 268
|
|
chr7_+_94536926
|
7.324
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr14_-_21493122
|
7.294
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr2_-_157189040
|
7.229
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr6_+_154407641
|
7.212
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr2_-_152955502
|
7.191
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr6_-_32095993
|
7.152
|
NM_001136153
NM_004381
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr5_-_79286793
|
7.064
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr13_+_113623496
|
7.029
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr8_-_82359620
|
7.011
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr5_-_141993691
|
7.011
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr14_+_92788924
|
7.007
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr8_+_42752019
|
6.991
|
NM_032410
|
HOOK3
|
hook homolog 3 (Drosophila)
|
|
chr10_+_101419169
|
6.978
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr3_-_49907304
|
6.865
|
NM_024046
|
CAMKV
|
CaM kinase-like vesicle-associated
|
|
chr16_-_30366681
|
6.849
|
NM_006110
|
CD2BP2
|
CD2 (cytoplasmic tail) binding protein 2
|
|
chr2_-_213403280
|
6.838
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr10_+_74451838
|
6.754
|
NM_138357
|
MCU
|
mitochondrial calcium uniporter
|
|
chr19_+_57831824
|
6.752
|
NM_213598
|
ZNF543
|
zinc finger protein 543
|
|
chr19_-_49944613
|
6.725
|
NM_020309
|
SLC17A7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7
|
|
chr19_-_4066739
|
6.723
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr17_-_36956145
|
6.693
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr13_-_67804432
|
6.689
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr22_+_39853316
|
6.661
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr7_-_17980102
|
6.652
|
NM_015132
|
SNX13
|
sorting nexin 13
|
|
chr4_+_154125589
|
6.588
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr10_-_120514669
|
6.570
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr10_-_118501981
|
6.510
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr12_+_50898744
|
6.502
|
NM_173602
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr2_-_176866925
|
6.476
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chr6_-_46293628
|
6.466
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr16_+_8814567
|
6.461
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr15_-_75744029
|
6.458
|
NM_001145358
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr2_-_86564632
|
6.403
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr3_+_51428698
|
6.385
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr19_-_38714781
|
6.266
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chrX_+_49687212
|
6.240
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr9_-_10612722
|
6.213
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr6_+_42018250
|
6.161
|
NM_138572
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr2_+_239335567
|
6.073
|
NM_001040445
|
ASB1
|
ankyrin repeat and SOCS box containing 1
|
|
chr2_+_128848752
|
6.046
|
NM_020120
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chrX_+_77166178
|
6.029
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr5_+_80597401
|
6.022
|
NM_001131035
NM_001131036
NM_032280
|
ZCCHC9
|
zinc finger, CCHC domain containing 9
|
|
chr15_-_83316621
|
6.019
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr17_-_15165873
|
6.018
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr1_-_161279724
|
6.012
|
NM_000530
|
MPZ
|
myelin protein zero
|
|
chr19_-_49149252
|
5.972
|
NM_001217
|
CA11
|
carbonic anhydrase XI
|
|
chr4_+_158141735
|
5.971
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr7_-_138665853
|
5.967
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr21_-_30257611
|
5.901
|
NM_013240
NM_182749
|
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative)
|
|
chr10_-_81205091
|
5.888
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr1_-_43205733
|
5.878
|
NM_001123395
NM_001185117
NM_148960
|
CLDN19
|
claudin 19
|
|
chr6_+_99282579
|
5.874
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr9_-_113800243
|
5.870
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr10_-_28571014
|
5.850
|
NM_173496
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7)
|
|
chr16_+_85061303
|
5.809
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr9_+_34958171
|
5.798
|
NM_015297
|
KIAA1045
|
KIAA1045
|
|
chr2_-_86565205
|
5.793
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr17_-_15164043
|
5.763
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr1_-_36235422
|
5.758
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr1_-_8483701
|
5.756
|
NM_001042682
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr8_-_27472298
|
5.751
|
NM_001831
|
CLU
|
clusterin
|
|
chr14_-_76448091
|
5.728
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chrX_+_68725077
|
5.718
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr12_+_51318515
|
5.711
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chr3_+_138666075
|
5.708
|
NM_001040061
|
C3orf72
|
chromosome 3 open reading frame 72
|
|
chr13_-_50510410
|
5.660
|
NM_001127482
NM_020456
|
SPRYD7
|
SPRY domain containing 7
|
|
chr8_-_87755880
|
5.656
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr4_-_7941092
|
5.632
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr12_+_54402795
|
5.625
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr15_+_51973549
|
5.610
|
NM_001165257
NM_013243
|
SCG3
|
secretogranin III
|
|
chr12_+_2162388
|
5.577
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr1_-_6240082
|
5.547
|
NM_015557
|
CHD5
|
chromodomain helicase DNA binding protein 5
|
|
chr2_-_217560247
|
5.530
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr16_-_20587625
|
5.529
|
NM_001105069
NM_182617
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B
|
|
chr17_-_71640226
|
5.467
|
NM_001144952
|
SDK2
|
sidekick cell adhesion molecule 2
|
|
chr16_-_49860917
|
5.466
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr8_+_19796581
|
5.440
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr1_+_33352071
|
5.430
|
NM_002143
|
HPCA
|
hippocalcin
|
|
chr11_-_102962903
|
5.407
|
NM_032299
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae)
|
|
chr6_+_36410543
|
5.379
|
NM_173562
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr19_-_49576120
|
5.367
|
NM_031886
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr2_+_183943286
|
5.359
|
NM_001142314
NM_080876
|
DUSP19
|
dual specificity phosphatase 19
|