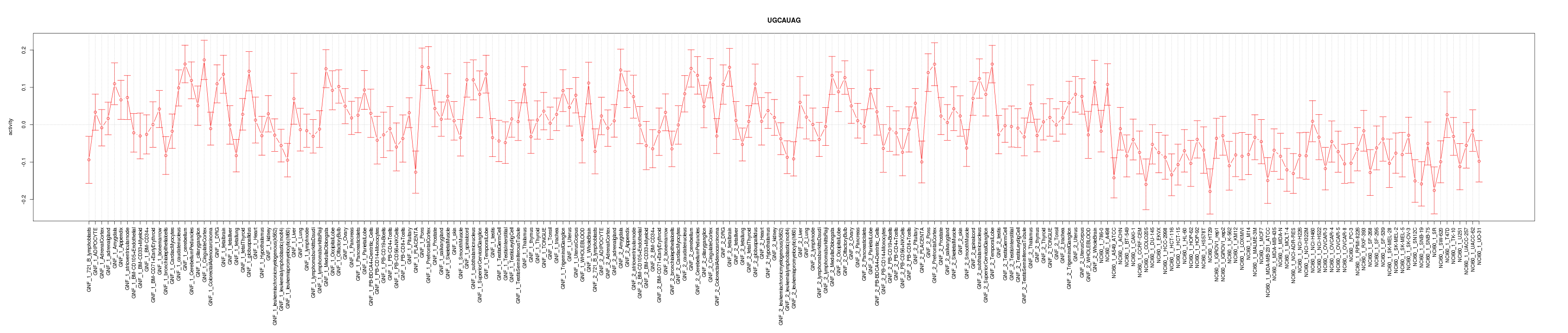
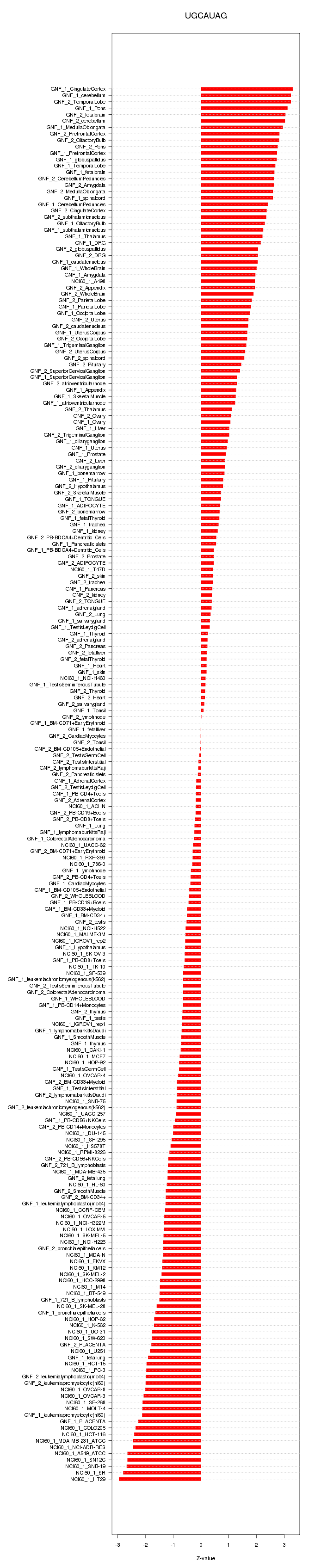
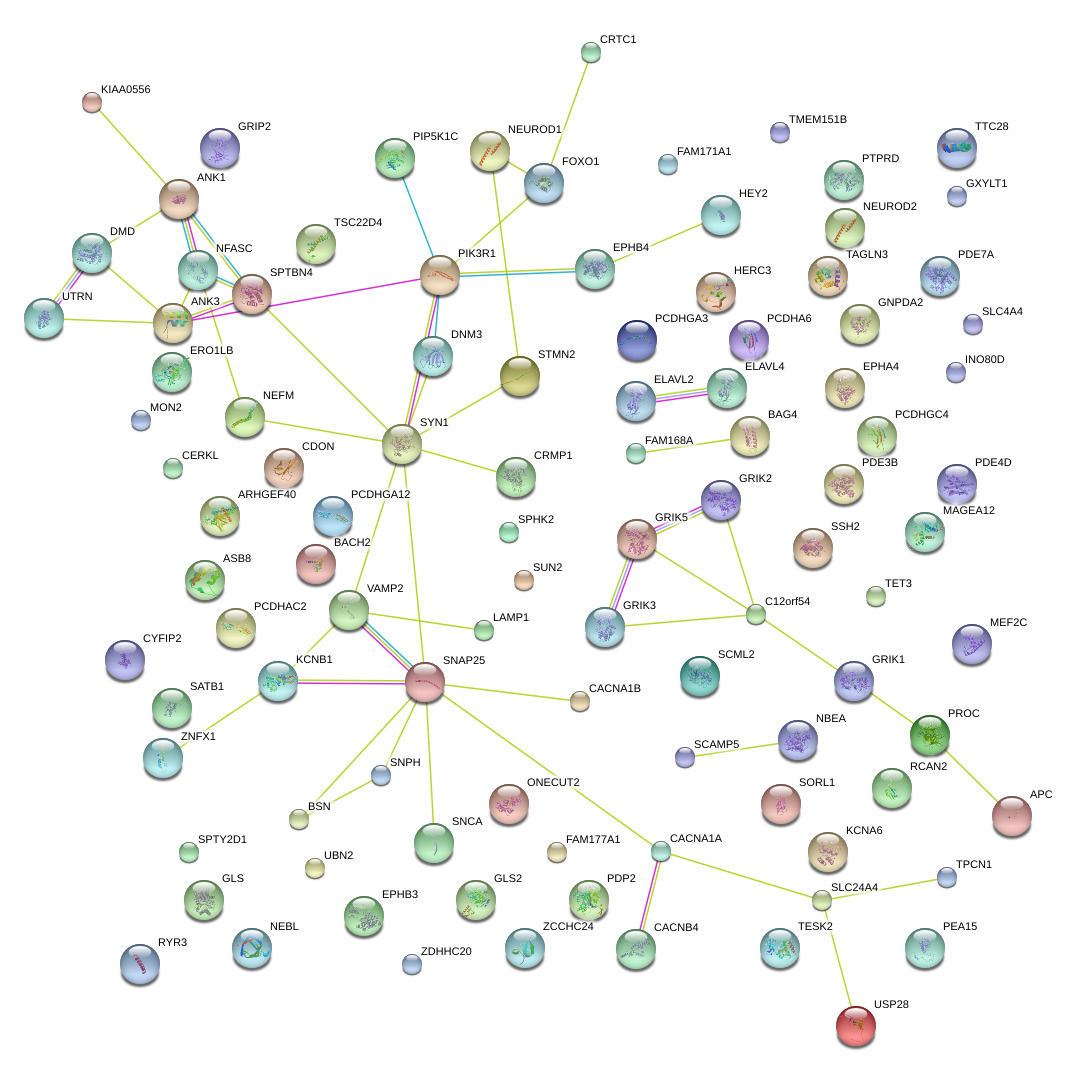
|
chr20_+_10199455
|
45.234
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr3_+_111717585
|
40.184
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr8_+_80523048
|
37.146
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr8_+_24771268
|
30.220
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr8_+_24772454
|
30.002
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr20_-_48099178
|
27.089
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr11_-_113746205
|
25.999
|
NM_020886
|
USP28
|
ubiquitin specific peptidase 28
|
|
chr17_-_8066254
|
24.623
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr1_+_204913353
|
24.424
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr1_+_171810611
|
24.018
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chrX_-_47479229
|
23.348
|
NM_006950
NM_133499
|
SYN1
|
synapsin I
|
|
chr3_+_111718006
|
22.989
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr17_-_37764165
|
22.079
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr3_+_49591897
|
21.550
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr6_-_46293628
|
20.322
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr4_-_44728518
|
20.149
|
NM_138335
|
GNPDA2
|
glucosamine-6-phosphate deaminase 2
|
|
chr4_-_90759446
|
20.105
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr11_-_18655962
|
19.802
|
NM_194285
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae)
|
|
chr8_+_38034066
|
19.575
|
NM_001204878
NM_004874
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr16_+_27561441
|
19.093
|
NM_015202
|
KIAA0556
|
KIAA0556
|
|
chr5_+_156693051
|
18.499
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr20_+_54933982
|
18.139
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr10_-_21463019
|
17.894
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr5_+_140864626
|
17.650
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr5_-_88179278
|
17.621
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_-_5894695
|
17.036
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr1_+_204797774
|
16.978
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr6_+_126070724
|
16.827
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr10_-_15413054
|
16.800
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr10_-_62149487
|
16.793
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr3_-_18480203
|
16.685
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr5_+_140739589
|
16.343
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr20_+_1246959
|
16.309
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr5_+_156696351
|
16.215
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr19_-_3700466
|
16.066
|
NM_001195733
NM_012398
|
PIP5K1C
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma
|
|
chr7_+_138916230
|
16.004
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr16_+_66914380
|
15.691
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr19_+_49122547
|
15.594
|
NM_001204158
NM_001243876
NM_020126
|
SPHK2
|
sphingosine kinase 2
|
|
chr10_-_62493282
|
15.535
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr2_-_152830448
|
15.394
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr4_-_90758126
|
15.346
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr12_+_4918341
|
15.270
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr19_+_49122789
|
15.245
|
NM_001204159
|
SPHK2
|
sphingosine kinase 2
|
|
chr14_+_35515347
|
14.961
|
NM_173607
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr5_+_140729723
|
14.857
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr3_-_18466759
|
14.694
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr14_+_92790151
|
14.665
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr4_-_5890266
|
14.660
|
NM_001313
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr5_+_67511578
|
14.288
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr11_-_73309090
|
14.250
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr11_+_121322848
|
14.069
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr5_+_140868721
|
14.068
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr5_+_67584251
|
13.972
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_-_91006460
|
13.964
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr5_-_58334936
|
13.871
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_-_66753968
|
13.705
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr10_-_81205091
|
13.626
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr5_+_140345746
|
13.463
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr5_+_140749830
|
13.394
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr12_+_113659216
|
13.188
|
NM_001143819
NM_017901
|
TPCN1
|
two pore segment channel 1
|
|
chr22_-_39152008
|
13.187
|
NM_001199579
NM_015374
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr6_-_91006580
|
12.815
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_-_152955208
|
12.699
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr5_+_140767402
|
12.291
|
NM_003736
NM_032098
|
PCDHGB4
|
protocadherin gamma subfamily B, 4
|
|
chr8_-_41754244
|
12.134
|
NM_001142446
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chrX_-_18372777
|
12.092
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr9_-_23821842
|
12.042
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr20_+_9494995
|
12.033
|
NM_001199897
NM_012261
|
LAMP5
|
lysosomal-associated membrane protein family, member 5
|
|
chr19_+_18794391
|
11.949
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr20_-_47894591
|
11.882
|
NM_021035
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr12_-_42538432
|
11.809
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr14_+_35514112
|
11.661
|
NM_001079519
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr5_+_140797243
|
11.643
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr14_+_92789536
|
11.576
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr2_-_222436996
|
11.301
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr12_-_48551269
|
11.298
|
NM_024095
|
ASB8
|
ankyrin repeat and SOCS box containing 8
|
|
chr6_+_101846860
|
11.290
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr22_-_29075708
|
11.278
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr2_-_152955502
|
11.244
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr7_-_100076754
|
11.241
|
NM_030935
|
TSC22D4
|
TSC22 domain family, member 4
|
|
chr6_+_44238479
|
11.093
|
NM_001137560
|
TMEM151B
|
transmembrane protein 151B
|
|
chr18_+_55102777
|
11.092
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr10_-_62332666
|
11.011
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr2_+_74273449
|
10.907
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr1_+_160175114
|
10.905
|
NM_003768
|
PEA15
|
phosphoprotein enriched in astrocytes 15
|
|
chr13_+_113951461
|
10.880
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr2_-_182545212
|
10.819
|
NM_002500
|
CERKL
NEUROD1
|
ceramide kinase-like
neurogenic differentiation 1
|
|
chr2_-_206950895
|
10.786
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr4_+_89513597
|
10.741
|
NM_014606
|
HERC3
|
hect domain and RLD 3
|
|
chr19_+_40973049
|
10.586
|
NM_020971
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chrX_-_32430370
|
10.563
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr5_+_112043188
|
10.517
|
NM_001127511
|
APC
|
adenomatous polyposis coli
|
|
chr13_+_35516390
|
10.446
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr15_+_75287829
|
10.325
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr9_-_10612722
|
10.189
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr5_+_140810126
|
10.170
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr15_+_33603162
|
9.991
|
NM_001036
NM_001243996
|
RYR3
|
ryanodine receptor 3
|
|
chr12_+_62860531
|
9.942
|
NM_015026
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr1_-_45956795
|
9.888
|
NM_007170
|
TESK2
|
testis-specific kinase 2
|
|
chr5_+_140753480
|
9.817
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr13_-_22033505
|
9.732
|
NM_153251
|
ZDHHC20
|
zinc finger, DHHC-type containing 20
|
|
chr5_+_140254930
|
9.653
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr5_+_140777603
|
9.650
|
NM_018925
NM_032099
|
PCDHGB5
|
protocadherin gamma subfamily B, 5
|
|
chr9_+_140772228
|
9.519
|
NM_000718
NM_001243812
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr14_+_21538339
|
9.519
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr4_+_72204769
|
9.431
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr17_-_28257017
|
9.395
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr5_-_58652671
|
9.367
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_72052354
|
9.341
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chrX_-_32173585
|
9.272
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr1_-_236445294
|
9.116
|
NM_019891
|
ERO1LB
|
ERO1-like beta (S. cerevisiae)
|
|
chr13_-_41240607
|
8.991
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr2_+_191745546
|
8.886
|
NM_014905
|
GLS
|
glutaminase
|
|
chr11_-_125933186
|
8.854
|
NM_001243597
NM_016952
|
CDON
|
Cdon homolog (mouse)
|
|
chr3_-_14583587
|
8.810
|
NM_001080423
|
GRIP2
|
glutamate receptor interacting protein 2
|
|
chr11_+_14665268
|
8.807
|
NM_000922
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr12_-_121736110
|
8.619
|
NM_006549
NM_153499
NM_153500
NM_172214
NM_172215
NM_172216
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr1_-_54872075
|
8.572
|
NM_001009955
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr5_+_140247767
|
8.539
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr5_+_140186658
|
8.524
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr9_-_23821477
|
8.452
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr6_-_150039391
|
8.360
|
NM_004690
|
LATS1
|
LATS, large tumor suppressor, homolog 1 (Drosophila)
|
|
chr3_-_10547267
|
8.352
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr5_+_140220811
|
8.351
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr1_+_41249479
|
8.351
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr20_+_48599511
|
8.236
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr5_+_140213968
|
8.177
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr11_+_26353677
|
8.126
|
NM_031418
|
ANO3
|
anoctamin 3
|
|
chr11_+_17757494
|
8.123
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr4_-_102268626
|
8.072
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr8_+_79578258
|
8.033
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chrX_+_135067573
|
8.006
|
NM_001177651
NM_001042537
NM_006359
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr4_-_25032306
|
7.996
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr5_-_58295758
|
7.972
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_140762401
|
7.934
|
NM_018920
NM_032087
|
PCDHGA7
|
protocadherin gamma subfamily A, 7
|
|
chr5_+_112073555
|
7.904
|
NM_000038
NM_001127510
|
APC
|
adenomatous polyposis coli
|
|
chr12_-_121734543
|
7.840
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr2_-_27485774
|
7.734
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr20_+_8112762
|
7.733
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr10_-_61900659
|
7.540
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr4_-_170947428
|
7.484
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr1_-_54871978
|
7.426
|
NM_018070
NM_145716
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chrX_+_10124984
|
7.410
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr14_+_92788924
|
7.365
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr7_+_39989955
|
7.338
|
NM_003718
NM_031267
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr2_-_11484699
|
7.269
|
NM_004850
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2
|
|
chr6_+_41040687
|
7.244
|
NM_002505
NM_021705
|
NFYA
|
nuclear transcription factor Y, alpha
|
|
chr5_+_139493689
|
7.163
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr15_-_78423556
|
7.144
|
NM_006383
|
CIB2
|
calcium and integrin binding family member 2
|
|
chr5_+_140771482
|
7.074
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chr7_+_45614081
|
7.071
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr2_+_26915390
|
7.062
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr9_-_23826062
|
7.008
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr20_-_43729607
|
6.936
|
NM_002251
|
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1
|
|
chrX_+_53111482
|
6.883
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr14_-_93799371
|
6.882
|
NM_001002860
NM_018167
|
BTBD7
|
BTB (POZ) domain containing 7
|
|
chr16_-_30022300
|
6.804
|
NM_003586
|
DOC2A
|
double C2-like domains, alpha
|
|
chrX_-_54384436
|
6.707
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chrX_-_33146543
|
6.705
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr12_+_78225068
|
6.698
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr3_+_155860750
|
6.658
|
NM_003471
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr22_-_39151457
|
6.655
|
NM_001199580
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr5_+_140734767
|
6.596
|
NM_018917
NM_032053
|
PCDHGA4
|
protocadherin gamma subfamily A, 4
|
|
chr11_-_85338313
|
6.588
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr19_-_1174263
|
6.552
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr12_-_122018858
|
6.450
|
NM_032590
|
KDM2B
|
lysine (K)-specific demethylase 2B
|
|
chr8_-_66701175
|
6.414
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr10_-_21186530
|
6.403
|
NM_006393
|
NEBL
|
nebulette
|
|
chr5_+_140306301
|
6.394
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr12_+_4382882
|
6.394
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr1_+_159141376
|
6.388
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr5_+_140800536
|
6.384
|
NM_018914
NM_032091
NM_032092
|
PCDHGA11
|
protocadherin gamma subfamily A, 11
|
|
chr5_+_140855543
|
6.241
|
NM_002588
NM_032402
NM_032403
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr2_+_45169036
|
6.214
|
NM_005413
|
SIX3
|
SIX homeobox 3
|
|
chr5_+_140718353
|
6.191
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr5_+_140165875
|
6.189
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr6_+_37787266
|
6.155
|
NM_021943
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr1_+_155051263
|
6.092
|
NM_004952
|
EFNA3
|
ephrin-A3
|
|
chr1_+_99729847
|
6.058
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr1_-_202679415
|
6.037
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr5_+_67588395
|
6.017
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr5_+_140743755
|
5.989
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr10_+_86088156
|
5.975
|
NM_018999
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr1_-_202612580
|
5.951
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr1_-_114354906
|
5.937
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr1_-_146644045
|
5.934
|
NM_005399
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit
|
|
chr3_+_178276487
|
5.924
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr3_-_66024508
|
5.889
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr5_-_59064437
|
5.887
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_+_101142754
|
5.877
|
NM_024708
NM_198243
|
ASB7
|
ankyrin repeat and SOCS box containing 7
|
|
chr7_+_43152196
|
5.819
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr12_+_32654976
|
5.788
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr15_+_31619043
|
5.737
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chrX_+_102883891
|
5.710
|
NM_001006639
NM_004780
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr9_+_112403067
|
5.704
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr12_+_5019048
|
5.676
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr1_+_52608045
|
5.567
|
NM_004799
NM_007323
NM_007324
|
ZFYVE9
|
zinc finger, FYVE domain containing 9
|
|
chr4_-_175750377
|
5.498
|
NM_001042543
NM_006529
|
GLRA3
|
glycine receptor, alpha 3
|
|
chr3_+_12045803
|
5.443
|
NM_003178
NM_133625
|
SYN2
|
synapsin II
|
|
chr16_+_8768403
|
5.435
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|