|
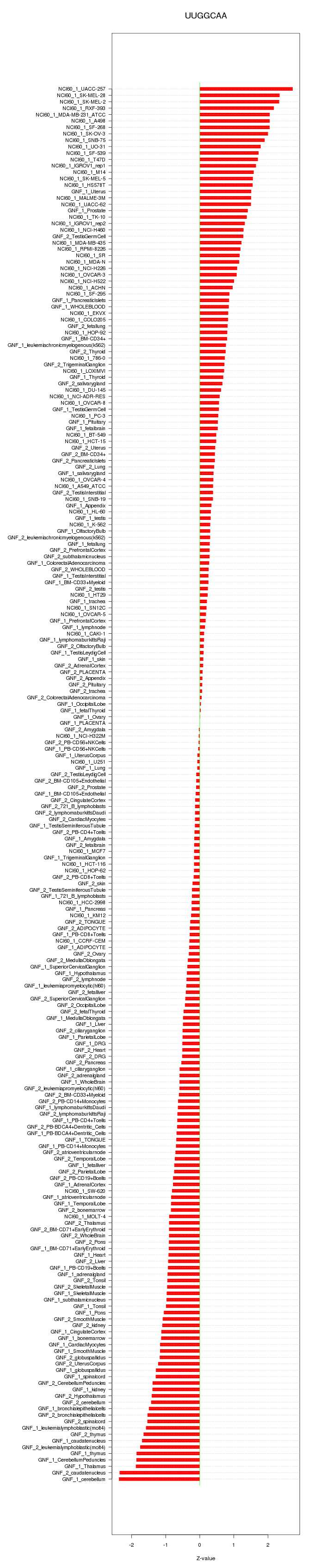
chr15_-_91537724
|
22.300
|
NM_003981
NM_199413
NM_199414
|
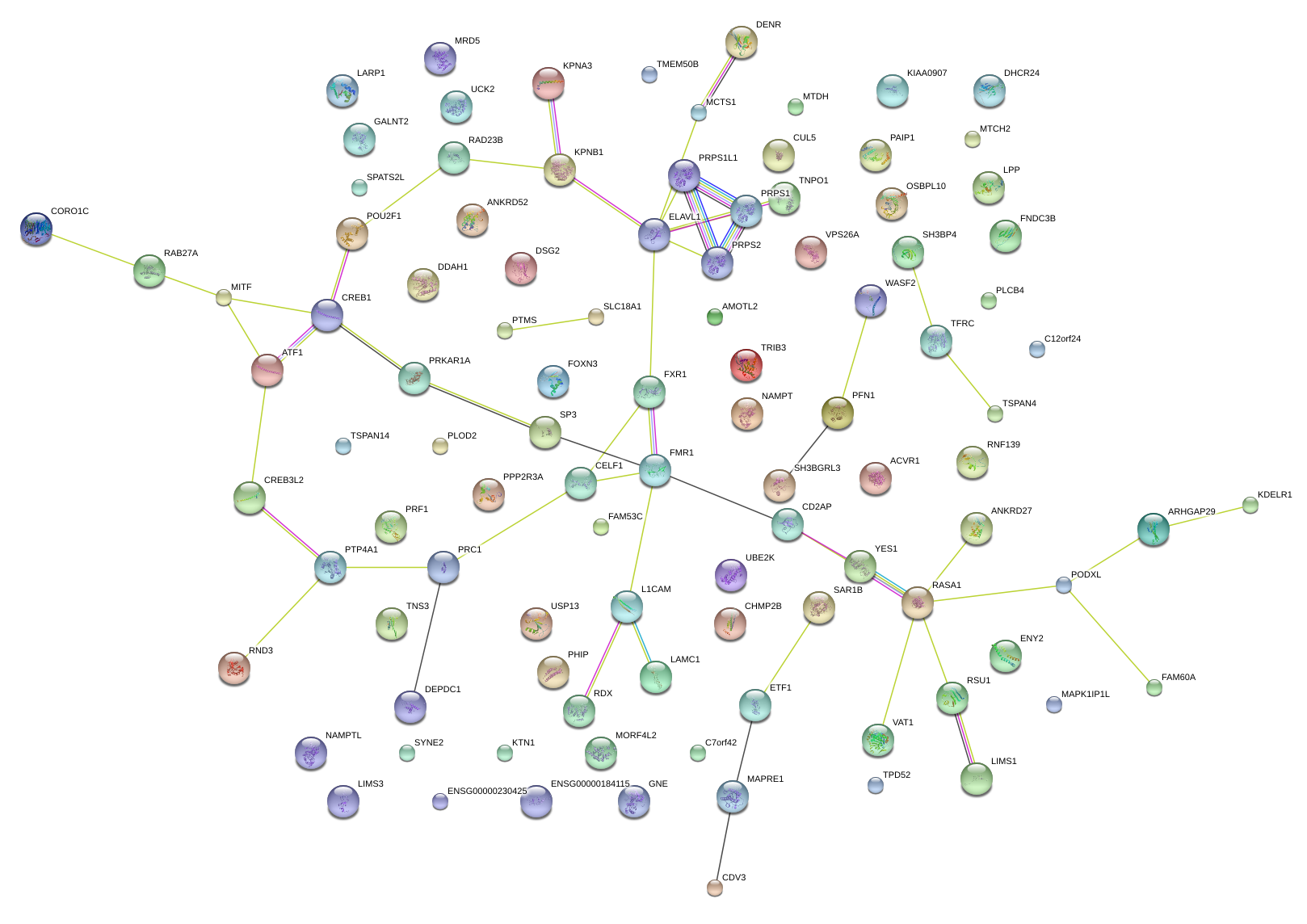
PRC1
|
protein regulator of cytokinesis 1
|
|
chr2_+_235860616
|
22.070
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr3_-_195808958
|
18.540
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr3_-_195808997
|
18.358
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr1_+_165796652
|
17.844
|
NM_012474
|
UCK2
|
uridine-cytidine kinase 2
|
|
chr2_+_109204904
|
16.878
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr3_+_180630054
|
16.777
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chrX_+_119737743
|
15.409
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr1_-_94703252
|
15.345
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chrX_+_119738542
|
15.344
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr2_+_109204745
|
15.089
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_+_109237679
|
14.955
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr19_-_48894755
|
14.597
|
NM_006801
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1
|
|
chr14_+_56046796
|
14.245
|
NM_001079521
NM_001079522
NM_004986
NM_182926
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr11_+_844062
|
14.158
|
NM_001025234
|
TSPAN4
|
tetraspanin 4
|
|
chr2_+_109223600
|
13.879
|
NM_001193482
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr18_-_812268
|
13.691
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr5_+_72112417
|
13.637
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr12_+_123237302
|
13.624
|
NM_003677
|
DENR
|
density-regulated protein
|
|
chr11_+_844445
|
13.509
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr3_+_187943192
|
13.499
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_+_69812620
|
13.469
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_-_145879222
|
13.388
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr6_+_47445440
|
13.323
|
NM_012120
|
CD2AP
|
CD2-associated protein
|
|
chr9_+_110045516
|
13.291
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr1_+_167298280
|
13.196
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr5_-_133968463
|
12.811
|
NM_001033503
NM_016103
|
SAR1B
|
SAR1 homolog B (S. cerevisiae)
|
|
chr7_-_137686814
|
12.662
|
NM_001253775
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr3_+_187930720
|
12.661
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr15_-_55562483
|
12.646
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr19_-_33166057
|
12.237
|
NM_032139
|
ANKRD27
|
ankyrin repeat domain 27 (VPS9 domain)
|
|
chr3_+_69812961
|
12.217
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr2_+_109271267
|
12.151
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_-_174828775
|
12.055
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr5_-_137878915
|
11.846
|
NM_004730
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr20_+_361272
|
11.709
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr2_-_158731622
|
11.479
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr7_-_47621741
|
11.420
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr3_-_134093235
|
11.311
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr12_-_109125289
|
11.233
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr5_+_72143929
|
11.222
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr11_+_842821
|
11.196
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr1_+_182992329
|
10.859
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr11_-_110167191
|
10.843
|
NM_002906
|
RDX
|
radixin
|
|
chr2_+_109150807
|
10.753
|
NM_001193483
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr17_+_45727274
|
10.725
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr13_-_50367027
|
10.719
|
NM_002267
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4)
|
|
chr4_+_39699663
|
10.516
|
NM_001111112
NM_001111113
NM_005339
|
UBE2K
|
ubiquitin-conjugating enzyme E2K
|
|
chrX_+_12809473
|
10.502
|
NM_001039091
NM_002765
|
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2
|
|
chr5_-_43557520
|
10.429
|
NM_183323
|
PAIP1
|
poly(A) binding protein interacting protein 1
|
|
chr3_+_69788562
|
10.385
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr17_-_4851797
|
10.307
|
NM_005022
|
PFN1
|
profilin 1
|
|
chr2_-_151344145
|
10.291
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr1_-_86043932
|
10.194
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr5_+_86564727
|
10.052
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr18_+_29078026
|
9.989
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr1_-_55352920
|
9.971
|
NM_014762
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr8_+_98656329
|
9.895
|
NM_178812
|
MTDH
|
metadherin
|
|
chr3_+_171757413
|
9.889
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr11_-_47664014
|
9.856
|
NM_014342
|
MTCH2
|
mitochondrial carrier 2
|
|
chr3_+_69915374
|
9.754
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_187871473
|
9.434
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr15_-_55563091
|
9.378
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr1_-_155904186
|
9.361
|
NM_014949
|
KIAA0907
|
KIAA0907
|
|
chr21_-_34852282
|
9.218
|
NM_006134
|
TMEM50B
|
transmembrane protein 50B
|
|
chr5_+_137673702
|
9.197
|
NM_016605
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr12_-_31479120
|
9.182
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chrX_-_102942968
|
9.160
|
NM_001142419
NM_001142420
NM_001142421
NM_001142422
NM_001142427
NM_001142428
NM_001142431
NM_001142432
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr1_-_27816676
|
9.103
|
NM_001201404
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr5_+_86564044
|
9.008
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr10_-_16859408
|
8.995
|
NM_012425
NM_152724
|
RSU1
|
Ras suppressor protein 1
|
|
chr2_+_201170795
|
8.983
|
NM_001100422
NM_001100424
NM_001100423
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chrX_-_153141398
|
8.942
|
NM_000425
NM_001143963
NM_024003
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr15_-_55581969
|
8.937
|
NM_183235
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr5_-_43557098
|
8.930
|
NM_006451
NM_182789
|
PAIP1
|
poly(A) binding protein interacting protein 1
|
|
chr2_-_174830429
|
8.928
|
NM_001172712
NM_003111
|
SP3
|
Sp3 transcription factor
|
|
chr8_-_80993009
|
8.893
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr7_-_131241321
|
8.853
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr5_+_137673223
|
8.845
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr3_+_133292404
|
8.819
|
NM_001134422
NM_017548
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr2_+_208394615
|
8.785
|
NM_004379
NM_134442
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr19_-_8070468
|
8.777
|
NM_001419
|
ELAVL1
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R)
|
|
chr11_-_47510518
|
8.666
|
NM_001025596
NM_001172640
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr1_+_230202928
|
8.552
|
NM_004481
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2)
|
|
chr17_+_66508525
|
8.515
|
NM_002734
NM_212472
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr10_+_82213892
|
8.513
|
NM_001128309
NM_030927
|
TSPAN14
|
tetraspanin 14
|
|
chr1_-_68962726
|
8.468
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr14_+_55518313
|
8.456
|
NM_144578
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr20_+_31407696
|
8.385
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr1_+_26606212
|
8.270
|
NM_031286
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr8_+_125486949
|
8.255
|
NM_007218
|
RNF139
|
ring finger protein 139
|
|
chr3_+_87276398
|
8.215
|
NM_001244644
NM_014043
|
CHMP2B
|
charged multivesicular body protein 2B
|
|
chr6_+_64281910
|
8.183
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chrX_+_146993450
|
8.179
|
NM_001185075
NM_001185076
NM_001185081
NM_001185082
NM_002024
|
FMR1
|
fragile X mental retardation 1
|
|
chr11_-_47516072
|
8.124
|
NM_198700
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr6_-_79787939
|
8.108
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr3_+_133293256
|
8.034
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr12_-_56652118
|
7.995
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr5_+_154092461
|
7.986
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr3_-_32023237
|
7.949
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr3_+_179370779
|
7.911
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr14_+_64680858
|
7.864
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr9_-_36276930
|
7.818
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr10_+_70883856
|
7.816
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr3_+_135741576
|
7.713
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr2_+_201170603
|
7.697
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr7_-_105925410
|
7.641
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr11_+_107879407
|
7.618
|
NM_003478
|
CUL5
|
cullin 5
|
|
chr3_+_171758289
|
7.501
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr7_+_66386129
|
7.483
|
NM_017994
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr12_+_110906203
|
7.309
|
NM_013300
|
C12orf24
|
chromosome 12 open reading frame 24
|
|
chr20_+_9288446
|
7.216
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr14_+_64682671
|
7.178
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr8_+_110346549
|
7.167
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr14_-_89883306
|
7.137
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr17_-_41174431
|
7.117
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr17_+_75446612
|
7.098
|
NM_001113496
|
SEPT9
|
septin 9
|
|
chr12_-_80307690
|
7.076
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr14_-_75643335
|
7.048
|
NM_006827
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr14_+_103801160
|
7.046
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr15_+_99192696
|
6.956
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr6_-_114292312
|
6.855
|
NM_001527
|
HDAC2
|
histone deacetylase 2
|
|
chr1_-_150669499
|
6.808
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr1_+_110546569
|
6.797
|
NM_001242675
NM_001242676
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr2_-_158732341
|
6.734
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr7_-_2281822
|
6.673
|
NM_013393
|
FTSJ2
|
FtsJ homolog 2 (E. coli)
|
|
chr17_+_66508098
|
6.609
|
NM_212471
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr7_+_17338182
|
6.589
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr20_+_9049660
|
6.546
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr3_+_38206968
|
6.469
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr8_+_56015013
|
6.409
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr12_-_76478737
|
6.395
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr1_+_89149836
|
6.268
|
NM_006256
|
PKN2
|
protein kinase N2
|
|
chr1_+_7831326
|
6.258
|
NM_004781
|
VAMP3
|
vesicle-associated membrane protein 3 (cellubrevin)
|
|
chr19_-_10514242
|
6.248
|
NM_007065
|
CDC37
|
cell division cycle 37 homolog (S. cerevisiae)
|
|
chr1_-_85930733
|
6.242
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr10_+_51576284
|
6.239
|
NM_001145262
|
NCOA4
|
nuclear receptor coactivator 4
|
|
chr17_-_37607314
|
6.195
|
NM_004774
|
MED1
|
mediator complex subunit 1
|
|
chr6_+_163835668
|
6.187
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr17_-_79828691
|
6.149
|
NM_001185077
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr2_+_85198176
|
6.126
|
NM_020122
|
KCMF1
|
potassium channel modulatory factor 1
|
|
chr10_+_114709963
|
6.117
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr9_-_36258383
|
6.063
|
NM_001190383
NM_001190384
NM_005476
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr16_-_87525317
|
6.061
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr12_-_49177876
|
5.981
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chr8_-_130951996
|
5.902
|
NM_016623
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr1_+_110543548
|
5.901
|
NM_001242674
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr1_-_153940187
|
5.878
|
NM_014437
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1
|
|
chr12_-_80329234
|
5.866
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr1_+_110091185
|
5.864
|
NM_006496
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr21_+_18885104
|
5.847
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr11_+_111895537
|
5.814
|
NM_001931
|
DLAT
|
dihydrolipoamide S-acetyltransferase
|
|
chr11_-_8680382
|
5.789
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chrX_+_49832214
|
5.719
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr20_+_9198036
|
5.700
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr11_-_47574766
|
5.693
|
NM_006560
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr17_+_75283972
|
5.681
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr9_+_115983807
|
5.661
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr1_+_201798281
|
5.659
|
NM_018085
|
IPO9
|
importin 9
|
|
chr10_+_62538088
|
5.629
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr17_+_34136437
|
5.610
|
NM_003487
NM_139215
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa
|
|
chr12_-_49182718
|
5.595
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr7_-_1595746
|
5.573
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr10_+_51565107
|
5.564
|
NM_001145260
NM_001145261
NM_001145263
|
NCOA4
|
nuclear receptor coactivator 4
|
|
chr3_-_57583153
|
5.541
|
NM_001660
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr6_-_109703703
|
5.451
|
NM_001142401
NM_001142402
NM_001142403
NM_001142404
NM_006016
|
CD164
|
CD164 molecule, sialomucin
|
|
chr2_-_47143250
|
5.396
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chrX_-_20284708
|
5.339
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr1_+_40723730
|
5.331
|
NM_005857
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr3_+_69985750
|
5.290
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr11_-_115375065
|
5.275
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr8_-_57906423
|
5.259
|
NM_017813
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr7_+_129710336
|
5.257
|
NM_014997
|
KLHDC10
|
kelch domain containing 10
|
|
chr10_-_14372807
|
5.248
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr2_+_169312793
|
5.231
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chr6_-_86352635
|
5.219
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr3_+_37903643
|
5.216
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr14_+_105331581
|
5.186
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr2_-_47142950
|
5.086
|
NM_001171506
NM_001171509
NM_139279
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr15_+_44829265
|
5.037
|
NM_003758
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr17_+_37617688
|
5.030
|
NM_015083
NM_016507
|
CDK12
|
cyclin-dependent kinase 12
|
|
chr19_+_34663311
|
5.010
|
NM_001114093
NM_015578
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr1_+_212208918
|
5.004
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr6_-_86352564
|
4.987
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr7_+_94285620
|
4.986
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr8_+_67687415
|
4.963
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr10_+_51572363
|
4.958
|
NM_005437
|
NCOA4
|
nuclear receptor coactivator 4
|
|
chr1_-_150947456
|
4.954
|
NM_022075
NM_181746
|
CERS2
|
ceramide synthase 2
|
|
chr22_+_45560529
|
4.919
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr20_-_48770302
|
4.891
|
NM_001162505
NM_199129
NM_199203
|
TMEM189
TMEM189-UBE2V1
|
transmembrane protein 189
TMEM189-UBE2V1 readthrough
|
|
chr6_-_86351156
|
4.882
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr10_+_75757847
|
4.878
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr6_+_88182624
|
4.808
|
NM_001168398
NM_006416
|
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1
|
|
chr14_-_75179739
|
4.791
|
NM_001039479
|
KIAA0317
|
KIAA0317
|
|
chr21_-_15755439
|
4.771
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr17_+_75471324
|
4.729
|
NM_001113495
|
SEPT9
|
septin 9
|
|
chr2_-_216300770
|
4.656
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr1_-_95392501
|
4.650
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr9_+_137533643
|
4.614
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr3_-_182698291
|
4.552
|
NM_020640
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae)
|