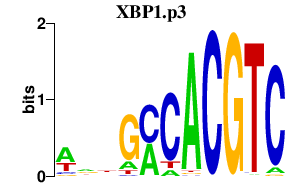
|
chr3_+_171758289
|
148.665
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr3_-_57583080
|
129.257
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr7_-_6523689
|
108.154
|
NM_001100603
NM_006854
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr3_-_57582866
|
103.965
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr3_-_57583090
|
101.055
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr2_-_69614321
|
99.375
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr6_-_7910716
|
96.791
|
NM_030810
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr1_-_43736600
|
95.112
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr6_-_7910278
|
94.470
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr3_-_57583153
|
90.534
|
NM_001660
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr7_+_128379345
|
88.452
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr15_+_79603427
|
87.183
|
NM_007364
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr15_+_79603497
|
86.420
|
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr3_+_133524671
|
80.540
|
|
SRPRB
|
signal recognition particle receptor, B subunit
|
|
chr15_-_60690115
|
79.251
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr3_+_133524636
|
76.326
|
|
SRPRB
|
signal recognition particle receptor, B subunit
|
|
chr12_-_106641675
|
74.922
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr3_+_127771287
|
73.237
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr7_+_128379400
|
69.663
|
|
CALU
|
calumenin
|
|
chr7_-_54826864
|
68.085
|
|
SEC61G
|
Sec61 gamma subunit
|
|
chr7_+_128379428
|
67.323
|
|
CALU
|
calumenin
|
|
chr7_+_128379452
|
67.220
|
|
CALU
|
calumenin
|
|
chr7_+_128379395
|
66.641
|
|
CALU
|
calumenin
|
|
chr1_-_24126031
|
66.119
|
NM_001127621
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr2_+_171785624
|
60.017
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr5_-_10761344
|
57.758
|
NM_004394
|
DAP
|
death-associated protein
|
|
chr11_-_66056607
|
56.783
|
NM_020470
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr5_-_131563481
|
55.116
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr2_+_171785780
|
54.987
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr3_+_100428393
|
54.896
|
|
TFG
|
TRK-fused gene
|
|
chr5_-_10761341
|
54.534
|
|
DAP
|
death-associated protein
|
|
chr7_+_94023872
|
54.170
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr11_+_17298278
|
53.836
|
NM_005013
|
NUCB2
|
nucleobindin 2
|
|
chr11_-_66056455
|
51.684
|
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr3_-_128369525
|
51.045
|
|
RPN1
|
ribophorin I
|
|
chr5_-_10761179
|
50.924
|
|
DAP
|
death-associated protein
|
|
chr2_-_20251788
|
50.507
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr12_+_124069053
|
50.468
|
NM_006815
|
TMED2
|
transmembrane emp24 domain trafficking protein 2
|
|
chr5_-_121414011
|
49.159
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr5_+_115177616
|
48.136
|
NM_001284
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit
|
|
chr2_-_33824294
|
48.100
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr11_+_61560352
|
47.833
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr14_-_24664758
|
47.398
|
NM_001014842
NM_006405
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr14_-_24664606
|
47.046
|
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr6_-_108279307
|
45.959
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr6_-_108279320
|
45.513
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr3_+_100428133
|
44.716
|
NM_001007565
NM_001195479
NM_006070
|
TFG
|
TRK-fused gene
|
|
chr16_-_75529278
|
44.353
|
|
|
|
|
chr10_+_89419352
|
43.972
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr11_+_61560108
|
43.808
|
NM_004111
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr14_-_39572414
|
43.324
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr12_+_124069099
|
42.806
|
|
TMED2
|
transmembrane emp24 domain trafficking protein 2
|
|
chr11_+_32112414
|
42.097
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr5_-_137088952
|
41.537
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr9_+_100818941
|
41.425
|
NM_018946
|
NANS
|
N-acetylneuraminic acid synthase
|
|
chr2_-_242212244
|
41.203
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr1_+_161123755
|
41.143
|
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1
|
|
chr21_-_18985104
|
40.952
|
|
BTG3
|
BTG family, member 3
|
|
chr7_-_54826899
|
40.823
|
NM_001012456
NM_014302
|
SEC61G
|
Sec61 gamma subunit
|
|
chr11_-_118927872
|
40.808
|
NM_006389
NM_001130991
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr6_-_109703553
|
40.056
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chrX_+_153060073
|
39.754
|
|
SSR4
|
signal sequence receptor, delta
|
|
chr11_+_69455938
|
39.225
|
|
CCND1
|
cyclin D1
|
|
chr11_-_61560052
|
38.876
|
NM_014206
|
C11orf10
|
chromosome 11 open reading frame 10
|
|
chr4_+_38869346
|
38.531
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr12_+_124069080
|
38.426
|
|
TMED2
|
transmembrane emp24 domain trafficking protein 2
|
|
chr8_-_124054491
|
38.368
|
|
DERL1
|
Der1-like domain family, member 1
|
|
chr14_+_52456322
|
38.270
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr11_-_118927851
|
37.234
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr1_-_11120001
|
37.109
|
|
SRM
|
spermidine synthase
|
|
chr17_-_40575117
|
36.992
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr17_+_15848230
|
36.751
|
NM_000676
|
ADORA2B
|
adenosine A2b receptor
|
|
chr20_+_44044706
|
36.133
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr21_-_18985224
|
36.116
|
NM_001130914
NM_006806
|
BTG3
|
BTG family, member 3
|
|
chr14_+_52456192
|
35.738
|
NM_016039
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr18_+_34409058
|
35.221
|
NM_020776
|
KIAA1328
|
KIAA1328
|
|
chr14_-_50087326
|
35.165
|
NM_001001
|
RPL36AL
|
ribosomal protein L36a-like
|
|
chr1_-_87379784
|
34.254
|
|
SEP15
|
15 kDa selenoprotein
|
|
chr1_-_87379808
|
33.901
|
|
SEP15
|
15 kDa selenoprotein
|
|
chr11_+_32605337
|
33.575
|
|
EIF3M
|
eukaryotic translation initiation factor 3, subunit M
|
|
chrX_+_153059903
|
33.249
|
NM_001204526
NM_006280
|
SSR4
|
signal sequence receptor, delta
|
|
chr2_+_85132762
|
32.984
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr10_-_120840296
|
32.963
|
|
EIF3A
|
eukaryotic translation initiation factor 3, subunit A
|
|
chr21_-_18985189
|
32.482
|
|
BTG3
|
BTG family, member 3
|
|
chr3_+_33155447
|
32.131
|
NM_006371
|
CRTAP
|
cartilage associated protein
|
|
chr3_+_100428361
|
32.077
|
NM_001195478
|
TFG
|
TRK-fused gene
|
|
chrX_+_153059629
|
32.058
|
NM_001204527
|
SSR4
|
signal sequence receptor, delta
|
|
chr1_-_11120076
|
31.833
|
|
SRM
|
spermidine synthase
|
|
chr11_+_32112607
|
31.697
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr6_+_33168602
|
31.608
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr20_+_31407696
|
31.579
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr3_+_122785855
|
31.549
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr11_-_118927815
|
31.413
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr7_-_30066124
|
31.262
|
NM_017946
|
FKBP14
|
FK506 binding protein 14, 22 kDa
|
|
chr1_-_11120081
|
31.045
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr1_-_45988561
|
30.839
|
NM_001202431
|
PRDX1
|
peroxiredoxin 1
|
|
chr10_-_71930185
|
30.418
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr8_-_124054540
|
29.533
|
|
DERL1
|
Der1-like domain family, member 1
|
|
chr1_-_11120047
|
29.478
|
|
SRM
|
spermidine synthase
|
|
chr13_-_31040035
|
29.050
|
NM_002128
|
HMGB1
|
high mobility group box 1
|
|
chr11_+_118443140
|
28.967
|
|
ARCN1
|
archain 1
|
|
chr1_+_228270429
|
28.788
|
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr4_-_119757183
|
28.722
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr9_-_35732226
|
28.449
|
|
TLN1
|
talin 1
|
|
chr2_+_85132999
|
28.414
|
|
TMSB10
|
thymosin beta 10
|
|
chr2_+_242254722
|
28.109
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr11_+_61560401
|
28.024
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr19_-_29704021
|
27.619
|
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
|
|
chr2_-_20251400
|
27.538
|
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr1_+_110527327
|
27.465
|
NM_001242673
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr8_-_124054591
|
27.228
|
NM_001134671
NM_024295
|
DERL1
|
Der1-like domain family, member 1
|
|
chrX_+_67718868
|
26.888
|
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr10_-_98346594
|
26.812
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr9_+_114393543
|
26.513
|
NM_001015882
NM_004125
|
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25
DNAJC25-GNG10 readthrough
|
|
chr11_-_62414086
|
26.485
|
|
GANAB
|
glucosidase, alpha; neutral AB
|
|
chr18_-_47018842
|
26.353
|
NM_001199356
NM_000985
NM_001035006
NM_001199340
NM_001199345
|
RPL17-C18ORF32
RPL17
|
RPL17-C18orf32 readthrough
ribosomal protein L17
|
|
chr2_-_242254984
|
26.215
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr6_-_90062349
|
26.141
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr6_+_30294224
|
26.040
|
|
|
|
|
chr18_-_47018777
|
25.954
|
|
RPL17
|
ribosomal protein L17
|
|
chr6_+_116692098
|
25.824
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr18_-_47018822
|
25.752
|
NM_001199341
NM_001199342
NM_001199343
NM_001199344
|
RPL17
|
ribosomal protein L17
|
|
chr1_+_87170486
|
25.604
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr17_+_15848422
|
25.558
|
|
ADORA2B
|
adenosine A2b receptor
|
|
chr21_+_42539727
|
25.497
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr13_-_31039966
|
25.044
|
|
HMGB1
|
high mobility group box 1
|
|
chr11_-_118972529
|
24.975
|
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase)
|
|
chr11_+_118443091
|
24.768
|
NM_001142281
NM_001655
|
ARCN1
|
archain 1
|
|
chr4_-_119757288
|
24.651
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr19_-_29704107
|
24.352
|
NM_006003
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
|
|
chr15_+_44038483
|
24.217
|
NM_005313
|
PDIA3
|
protein disulfide isomerase family A, member 3
|
|
chr11_-_62414063
|
24.001
|
|
GANAB
|
glucosidase, alpha; neutral AB
|
|
chr15_+_44038649
|
23.607
|
|
PDIA3
|
protein disulfide isomerase family A, member 3
|
|
chr11_-_62414096
|
23.456
|
NM_198334
NM_198335
|
GANAB
|
glucosidase, alpha; neutral AB
|
|
chr17_+_37026309
|
23.408
|
|
LASP1
|
LIM and SH3 protein 1
|
|
chrX_+_77359665
|
23.127
|
NM_000291
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr2_-_20251280
|
23.010
|
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr2_+_216176808
|
22.803
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr10_-_98346793
|
22.797
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_-_87380008
|
22.706
|
NM_004261
NM_203341
|
SEP15
|
15 kDa selenoprotein
|
|
chr5_-_121413906
|
22.411
|
|
LOX
|
lysyl oxidase
|
|
chrX_+_77359768
|
22.012
|
|
PGK1
|
phosphoglycerate kinase 1
|
|
chrX_+_77359754
|
21.931
|
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr12_+_58120029
|
21.656
|
|
LOC100130776
|
uncharacterized LOC100130776
|
|
chr22_-_43253186
|
21.632
|
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr12_-_101801499
|
21.528
|
NM_001177
|
ARL1
|
ADP-ribosylation factor-like 1
|
|
chr1_+_110527432
|
21.338
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr2_-_99952779
|
20.887
|
NM_005783
|
TXNDC9
|
thioredoxin domain containing 9
|
|
chr3_-_49395751
|
20.532
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr9_-_35732175
|
20.422
|
|
TLN1
|
talin 1
|
|
chr15_-_90645621
|
19.989
|
|
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial
|
|
chr17_+_37026351
|
19.519
|
|
LASP1
|
LIM and SH3 protein 1
|
|
chr9_+_91926109
|
19.502
|
NM_001827
|
CKS2
|
CDC28 protein kinase regulatory subunit 2
|
|
chrX_+_77359712
|
19.405
|
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr12_-_109125289
|
19.393
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr9_-_35072545
|
19.367
|
|
VCP
|
valosin containing protein
|
|
chr9_+_35732629
|
19.281
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr7_+_94024201
|
19.084
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr6_-_90062439
|
19.034
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr2_+_198365113
|
18.969
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr19_-_2038937
|
18.852
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr8_-_81083660
|
18.522
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr11_+_125462751
|
18.503
|
|
STT3A
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr9_+_35732498
|
18.421
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr4_-_47916498
|
18.205
|
NM_152995
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1
|
|
chr15_+_44038659
|
18.026
|
|
PDIA3
|
protein disulfide isomerase family A, member 3
|
|
chrX_+_47050208
|
18.000
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr1_+_110527307
|
17.468
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr12_+_54718927
|
17.233
|
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr6_-_109702902
|
17.217
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr9_-_35732115
|
17.132
|
|
TLN1
|
talin 1
|
|
chr1_+_87170515
|
17.097
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr22_-_43253304
|
17.038
|
NM_001142293
NM_014570
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr6_+_34204663
|
17.004
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr3_-_10362724
|
16.951
|
NM_001136232
|
SEC13
|
SEC13 homolog (S. cerevisiae)
|
|
chr2_+_200819928
|
16.757
|
NM_024520
|
C2orf47
|
chromosome 2 open reading frame 47
|
|
chr12_+_54718910
|
16.433
|
NM_016057
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr12_-_92539614
|
16.350
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr12_+_54718925
|
16.279
|
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr2_+_27255745
|
16.244
|
NM_001083590
NM_017727
|
TMEM214
|
transmembrane protein 214
|
|
chr2_+_27255837
|
16.230
|
|
TMEM214
|
transmembrane protein 214
|
|
chr6_+_7107986
|
16.132
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr12_-_82752154
|
15.727
|
NM_014167
|
CCDC59
|
coiled-coil domain containing 59
|
|
chr2_-_242212198
|
15.705
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr5_+_34656367
|
15.524
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr7_-_128045995
|
15.345
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr14_-_39572278
|
15.250
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr11_+_65819803
|
15.229
|
NM_006842
|
SF3B2
|
splicing factor 3b, subunit 2, 145kDa
|
|
chr10_-_16859336
|
15.040
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr9_+_35732207
|
15.016
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr16_-_22385596
|
14.980
|
NM_001802
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa
|
|
chr6_+_34204690
|
14.745
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr20_-_17662828
|
14.633
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr15_-_90645709
|
14.586
|
|
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial
|
|
chr12_+_57623827
|
14.394
|
NM_001166358
NM_001166359
NM_001166357
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial)
|
|
chr5_+_149340287
|
14.379
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr5_-_9546157
|
14.368
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr16_+_66586479
|
14.057
|
|
CKLF
|
chemokine-like factor
|
|
chr19_+_54618789
|
14.028
|
NM_015629
|
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr3_+_127771126
|
13.921
|
NM_013336
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|