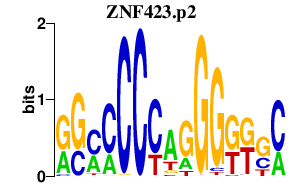
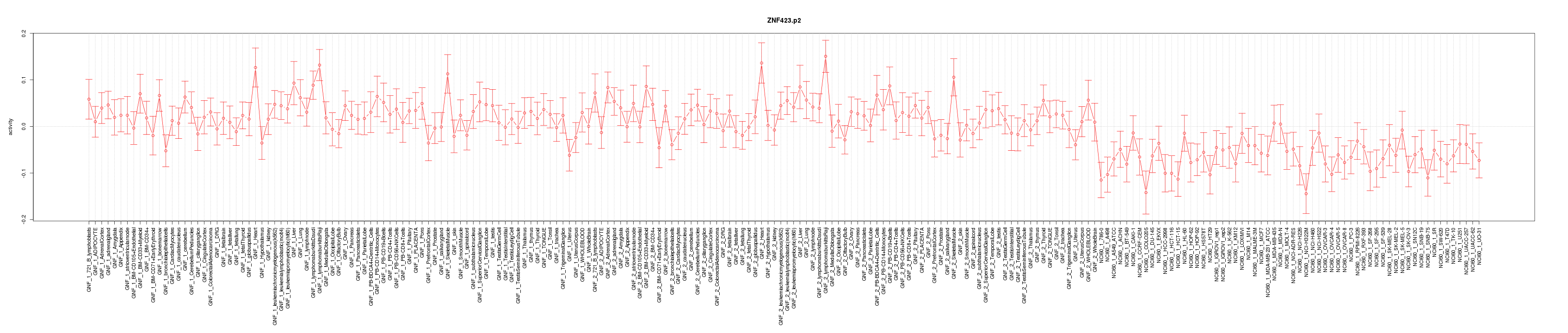
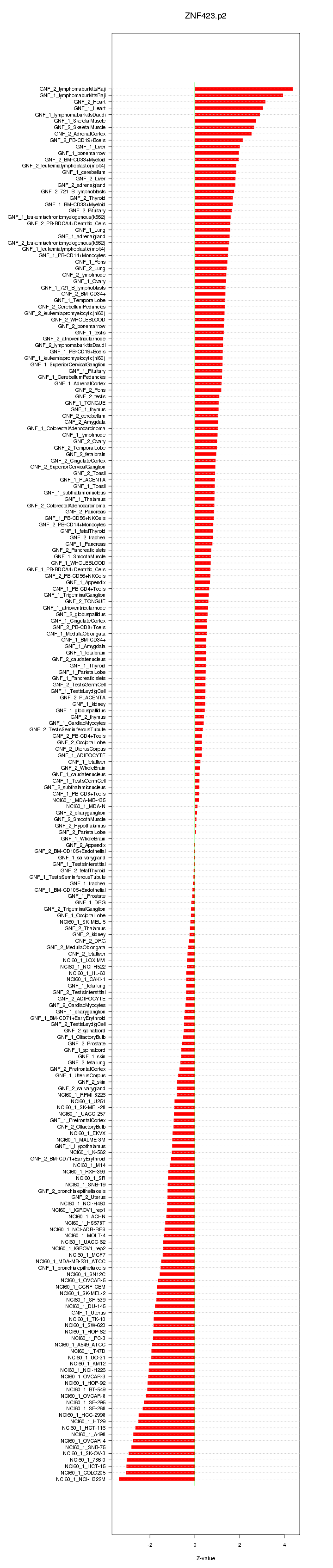
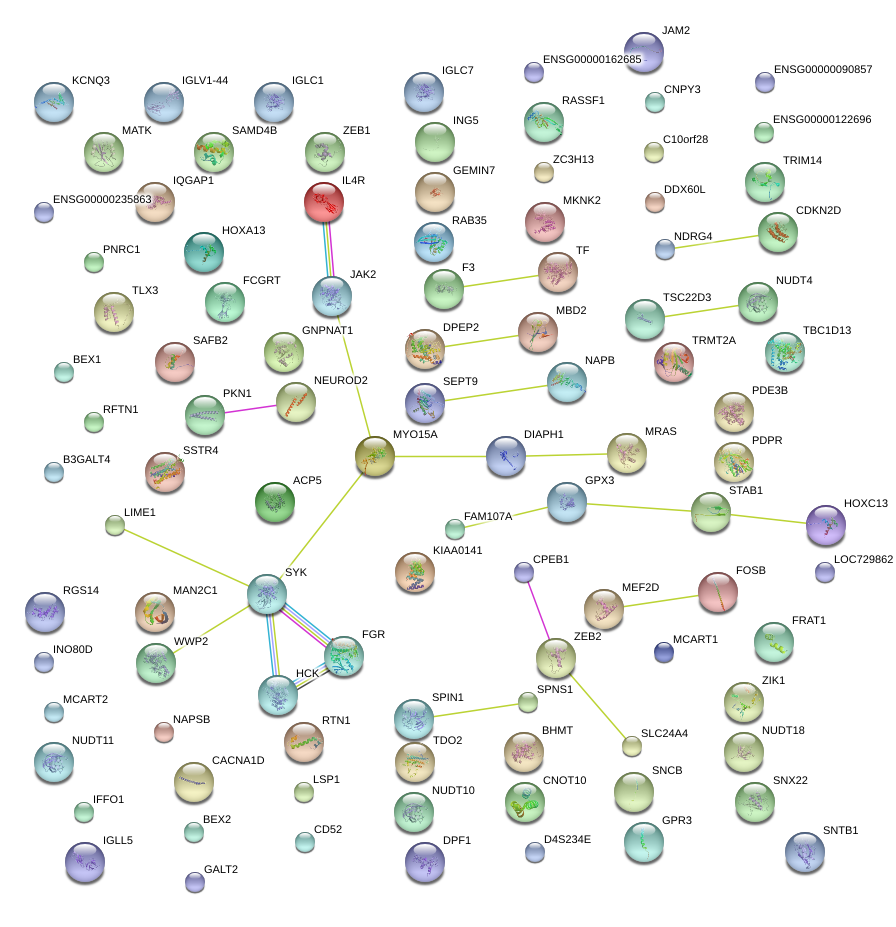
|
chr1_-_27952591
|
22.541
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chrX_-_107019001
|
17.698
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr14_+_92790151
|
17.334
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr19_+_45971249
|
16.861
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr1_+_26644458
|
15.928
|
|
CD52
|
CD52 molecule
|
|
chr10_+_99079017
|
15.701
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr5_+_150400132
|
14.052
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr9_+_93564063
|
13.987
|
|
SYK
|
spleen tyrosine kinase
|
|
chr9_+_93564099
|
13.920
|
|
SYK
|
spleen tyrosine kinase
|
|
chr3_+_52529351
|
13.549
|
NM_015136
|
STAB1
|
stabilin 1
|
|
chr3_+_52529370
|
13.521
|
|
STAB1
|
stabilin 1
|
|
chr9_+_93564073
|
13.474
|
|
SYK
|
spleen tyrosine kinase
|
|
chr5_+_150399980
|
13.210
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr16_+_70148213
|
12.494
|
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr9_+_93564204
|
12.020
|
NM_001174168
|
SYK
|
spleen tyrosine kinase
|
|
chr12_+_54332575
|
11.939
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr19_-_2041862
|
11.914
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chrX_-_102319080
|
11.899
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr2_-_145274916
|
11.682
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr8_-_133492556
|
11.557
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr19_-_3786279
|
11.458
|
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr11_+_1892098
|
11.371
|
NM_001013254
NM_001013255
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr3_+_138067969
|
11.363
|
NM_001252090
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr17_-_37764165
|
11.078
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr5_+_170736287
|
10.815
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr16_-_74401991
|
10.574
|
|
LOC283922
|
pyruvate dehydrogenase phosphatase regulatory subunit pseudogene
|
|
chr1_+_26644380
|
10.546
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr8_-_121823696
|
10.541
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chrX_-_51239426
|
10.247
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr19_-_2042022
|
10.221
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr6_+_89791510
|
10.216
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr19_-_5622794
|
9.950
|
NM_014649
|
SAFB2
|
scaffold attachment factor B2
|
|
chr9_+_93564011
|
9.693
|
NM_001135052
NM_003177
|
SYK
|
spleen tyrosine kinase
|
|
chr15_-_75660944
|
9.649
|
|
MAN2C1
|
mannosidase, alpha, class 2C, member 1
|
|
chr19_+_50016500
|
9.628
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr3_+_133465250
|
9.596
|
|
TF
|
transferrin
|
|
chr1_-_156470506
|
9.526
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr22_+_22786283
|
9.379
|
|
|
|
|
chr9_-_100881515
|
9.297
|
|
TRIM14
|
tripartite motif containing 14
|
|
chr13_-_46626845
|
9.273
|
NM_015070
|
ZC3H13
|
zinc finger CCCH-type containing 13
|
|
chr3_+_32726659
|
9.221
|
NM_015442
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10
|
|
chr19_+_50016490
|
9.091
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr3_+_133465204
|
9.091
|
|
TF
|
transferrin
|
|
chr6_+_33244916
|
9.048
|
NM_003782
|
B3GALT4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4
|
|
chr15_+_90931509
|
8.972
|
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr19_+_58095507
|
8.626
|
NM_001010879
|
ZIK1
|
zinc finger protein interacting with K protein 1 homolog (mouse)
|
|
chr15_+_90931489
|
8.608
|
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr11_+_14665308
|
8.570
|
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr3_-_58563070
|
8.539
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr12_-_120554558
|
8.492
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr4_-_169401571
|
8.487
|
|
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like
|
|
chr11_+_14665338
|
8.486
|
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr8_-_133492751
|
8.404
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr6_+_42897184
|
8.376
|
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr20_+_30639981
|
8.341
|
NM_001172129
NM_001172130
NM_001172131
NM_001172132
NM_001172133
NM_002110
|
HCK
|
hemopoietic cell kinase
|
|
chr20_+_23015949
|
8.322
|
NM_001052
|
SSTR4
|
somatostatin receptor 4
|
|
chrX_-_107018889
|
8.075
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr14_-_60097314
|
7.982
|
|
RTN1
|
reticulon 1
|
|
chr2_-_206950895
|
7.820
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr5_+_141303366
|
7.805
|
NM_001142603
NM_014773
|
KIAA0141
|
KIAA0141
|
|
chr8_-_21966812
|
7.759
|
NM_024815
|
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18
|
|
chr19_+_39833096
|
7.754
|
|
SAMD4B
|
sterile alpha motif domain containing 4B
|
|
chr16_+_58498216
|
7.687
|
|
NDRG4
|
NDRG family member 4
|
|
chr12_-_120554551
|
7.645
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr5_-_140998480
|
7.577
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr12_-_6665220
|
7.496
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr19_-_38714781
|
7.472
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr22_+_22735134
|
7.439
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr10_+_99894426
|
7.422
|
|
C10orf28
|
chromosome 10 open reading frame 28
|
|
chr20_-_23402038
|
7.343
|
NM_022080
|
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta
|
|
chr15_-_83316621
|
7.271
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr15_+_64443909
|
7.241
|
NM_024798
|
SNX22
|
sorting nexin 22
|
|
chr12_-_120554453
|
7.233
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr9_-_37904098
|
7.221
|
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr19_+_45582469
|
7.174
|
NM_001007270
NM_001007269
NM_024707
|
GEMIN7
|
gem (nuclear organelle) associated protein 7
|
|
chr1_+_27719151
|
7.162
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr5_+_78407603
|
7.076
|
NM_001713
|
BHMT
|
betaine--homocysteine S-methyltransferase
|
|
chr3_-_50374749
|
7.026
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr3_-_50374795
|
6.905
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr1_-_156470538
|
6.871
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr22_-_20104745
|
6.848
|
NM_022727
NM_182984
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr3_-_16555128
|
6.808
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr12_-_120554557
|
6.806
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr5_-_176057324
|
6.772
|
NM_001001502
NM_003085
|
SNCB
|
synuclein, beta
|
|
chr5_+_176784843
|
6.765
|
NM_006480
|
RGS14
|
regulator of G-protein signaling 14
|
|
chrX_-_51239295
|
6.760
|
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr16_+_28986024
|
6.743
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr19_-_3786382
|
6.712
|
NM_139354
NM_139355
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr16_+_69796237
|
6.599
|
NM_007014
NM_199423
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr9_+_131549561
|
6.589
|
|
TBC1D13
|
TBC1 domain family, member 13
|
|
chr20_+_62368035
|
6.583
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr19_-_10679620
|
6.578
|
NM_001800
NM_079421
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr19_-_11689765
|
6.555
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr14_-_53258329
|
6.513
|
NM_198066
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr9_+_4985243
|
6.509
|
NM_004972
|
JAK2
|
Janus kinase 2
|
|
chr18_-_51750460
|
6.500
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr3_+_53529030
|
6.402
|
NM_000720
NM_001128839
NM_001128840
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr4_+_4388804
|
6.391
|
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr2_+_242641310
|
6.376
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr5_+_141303463
|
6.367
|
|
KIAA0141
|
KIAA0141
|
|
chr17_+_18012019
|
6.352
|
NM_016239
|
MYO15A
|
myosin XVA
|
|
chr16_+_27325244
|
6.345
|
NM_000418
NM_001008699
|
IL4R
|
interleukin 4 receptor
|
|
chr19_+_14544100
|
6.313
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr21_+_27012067
|
6.252
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr11_-_64684606
|
6.212
|
NM_015104
|
ATG2A
|
ATG2 autophagy related 2 homolog A (S. cerevisiae)
|
|
chr21_-_35831856
|
6.192
|
NM_001127668
NM_001127669
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr7_-_100254048
|
6.178
|
NM_016188
|
ACTL6B
|
actin-like 6B
|
|
chr16_-_56459302
|
6.170
|
|
AMFR
|
autocrine motility factor receptor
|
|
chr8_-_80679897
|
6.151
|
NM_001040708
NM_012258
|
HEY1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr15_+_25200069
|
6.147
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chrX_+_70315637
|
6.111
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr21_+_27011766
|
6.110
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr6_+_42897236
|
6.077
|
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr9_-_139094886
|
6.069
|
NM_014564
|
LHX3
|
LIM homeobox 3
|
|
chr1_+_236849753
|
6.048
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr4_+_4388717
|
6.020
|
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr3_-_16554992
|
6.008
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr19_-_57183122
|
5.995
|
NM_001005850
|
ZNF835
|
zinc finger protein 835
|
|
chr9_+_137967401
|
5.966
|
|
OLFM1
|
olfactomedin 1
|
|
chr11_+_17757494
|
5.949
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr14_+_103995535
|
5.784
|
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr11_+_12695851
|
5.748
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr11_+_14665268
|
5.732
|
NM_000922
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr1_+_110254863
|
5.692
|
NM_000851
|
GSTM5
|
glutathione S-transferase mu 5
|
|
chr11_+_819696
|
5.640
|
|
PNPLA2
|
patatin-like phospholipase domain containing 2
|
|
chr7_-_152373163
|
5.632
|
NM_005431
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2
|
|
chr8_-_133493003
|
5.631
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr3_+_6902812
|
5.612
|
|
GRM7
|
glutamate receptor, metabotropic 7
|
|
chr9_-_130742803
|
5.549
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr19_+_39833099
|
5.545
|
NM_018028
|
SAMD4B
|
sterile alpha motif domain containing 4B
|
|
chr3_-_50374875
|
5.535
|
NM_170713
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr20_-_23066828
|
5.520
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr9_-_91793375
|
5.503
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr20_+_44650260
|
5.487
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr19_-_11689591
|
5.486
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr14_+_103995473
|
5.465
|
NM_152307
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr12_-_4554779
|
5.428
|
NM_020996
|
FGF6
|
fibroblast growth factor 6
|
|
chr22_-_20104695
|
5.427
|
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr1_-_27953046
|
5.421
|
NM_001042729
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr1_-_212873197
|
5.379
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr8_-_144241918
|
5.362
|
NM_001130478
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr19_-_36523540
|
5.343
|
NM_001199570
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr16_+_230332
|
5.341
|
NM_005331
|
HBQ1
|
hemoglobin, theta 1
|
|
chr5_+_176784947
|
5.301
|
|
|
|
|
chr22_+_37257004
|
5.298
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr20_+_24929862
|
5.296
|
NM_003650
|
CST7
|
cystatin F (leukocystatin)
|
|
chr9_+_131549509
|
5.290
|
NM_018201
|
TBC1D13
|
TBC1 domain family, member 13
|
|
chr19_+_36359346
|
5.286
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr9_-_100881462
|
5.258
|
NM_014788
NM_033219
NM_033220
|
TRIM14
|
tripartite motif containing 14
|
|
chr9_-_37904331
|
5.193
|
NM_033412
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr3_+_137483133
|
5.167
|
NM_004189
|
SOX14
|
SRY (sex determining region Y)-box 14
|
|
chr21_+_45719964
|
5.152
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr18_-_5543967
|
5.146
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr21_+_47744032
|
5.144
|
NM_006031
|
PCNT
|
pericentrin
|
|
chr7_+_6629658
|
5.143
|
NM_024067
|
C7orf26
|
chromosome 7 open reading frame 26
|
|
chr19_-_5622725
|
5.140
|
|
SAFB2
|
scaffold attachment factor B2
|
|
chr16_-_56459371
|
5.123
|
NM_001144
|
AMFR
|
autocrine motility factor receptor
|
|
chrX_+_133507299
|
5.083
|
NM_001015877
NM_032335
NM_032458
|
PHF6
|
PHD finger protein 6
|
|
chr21_+_45719921
|
5.069
|
NM_002626
|
PFKL
|
phosphofructokinase, liver
|
|
chr15_-_79382839
|
5.050
|
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr14_-_53258145
|
5.044
|
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr19_+_22235278
|
5.031
|
|
ZNF257
|
zinc finger protein 257
|
|
chr6_-_32160289
|
5.008
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr19_-_15311773
|
4.992
|
NM_000435
|
NOTCH3
|
notch 3
|
|
chr9_+_4985082
|
4.978
|
|
JAK2
|
Janus kinase 2
|
|
chr10_+_99894380
|
4.929
|
NM_014472
|
C10orf28
|
chromosome 10 open reading frame 28
|
|
chr9_+_6757919
|
4.917
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr4_-_120549115
|
4.915
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr19_-_38806546
|
4.886
|
NM_001039672
NM_001039673
NM_001145463
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae)
|
|
chr4_+_76439653
|
4.857
|
NM_144721
|
THAP6
|
THAP domain containing 6
|
|
chr19_-_12777540
|
4.845
|
NM_000528
NM_001173498
|
MAN2B1
|
mannosidase, alpha, class 2B, member 1
|
|
chr14_-_60097531
|
4.841
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr19_-_14117096
|
4.820
|
NM_002918
|
RFX1
|
regulatory factor X, 1 (influences HLA class II expression)
|
|
chr16_-_1843652
|
4.810
|
NM_001146006
NM_004970
|
IGFALS
|
insulin-like growth factor binding protein, acid labile subunit
|
|
chr11_+_76883823
|
4.807
|
|
MYO7A
|
myosin VIIA
|
|
chr4_+_71570768
|
4.804
|
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chrX_+_152770083
|
4.797
|
|
BGN
|
biglycan
|
|
chr19_-_16653202
|
4.790
|
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein
|
|
chr7_-_100253976
|
4.769
|
|
|
|
|
chr10_+_88428263
|
4.762
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr22_+_22735207
|
4.761
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr19_+_45754551
|
4.730
|
|
MARK4
|
MAP/microtubule affinity-regulating kinase 4
|
|
chr17_+_77020278
|
4.718
|
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr19_+_55085237
|
4.708
|
NM_001130917
NM_006866
|
LILRA2
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2
|
|
chr19_+_22235253
|
4.699
|
NM_033468
|
ZNF257
|
zinc finger protein 257
|
|
chr15_-_75660905
|
4.685
|
NM_006715
|
MAN2C1
|
mannosidase, alpha, class 2C, member 1
|
|
chr19_-_46389307
|
4.683
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr8_+_121823538
|
4.681
|
|
|
|
|
chr3_-_9885684
|
4.622
|
|
RPUSD3
|
RNA pseudouridylate synthase domain containing 3
|
|
chr14_-_60097223
|
4.606
|
|
RTN1
|
reticulon 1
|
|
chr14_+_65381178
|
4.599
|
|
CHURC1-FNTB
CHURC1
|
CHURC1-FNTB readthrough
churchill domain containing 1
|
|
chr11_+_819563
|
4.596
|
|
PNPLA2
|
patatin-like phospholipase domain containing 2
|
|
chr21_+_27012045
|
4.588
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr6_+_292033
|
4.581
|
NM_020185
|
DUSP22
|
dual specificity phosphatase 22
|
|
chr1_-_183559463
|
4.561
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr11_-_1036654
|
4.551
|
NM_005961
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming
|
|
chr9_-_135230129
|
4.545
|
|
SETX
|
senataxin
|
|
chr16_+_71392615
|
4.544
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr8_-_145669796
|
4.535
|
NM_013432
|
TONSL
|
tonsoku-like, DNA repair protein
|
|
chr22_+_22735202
|
4.524
|
|
|
|