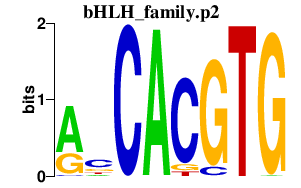
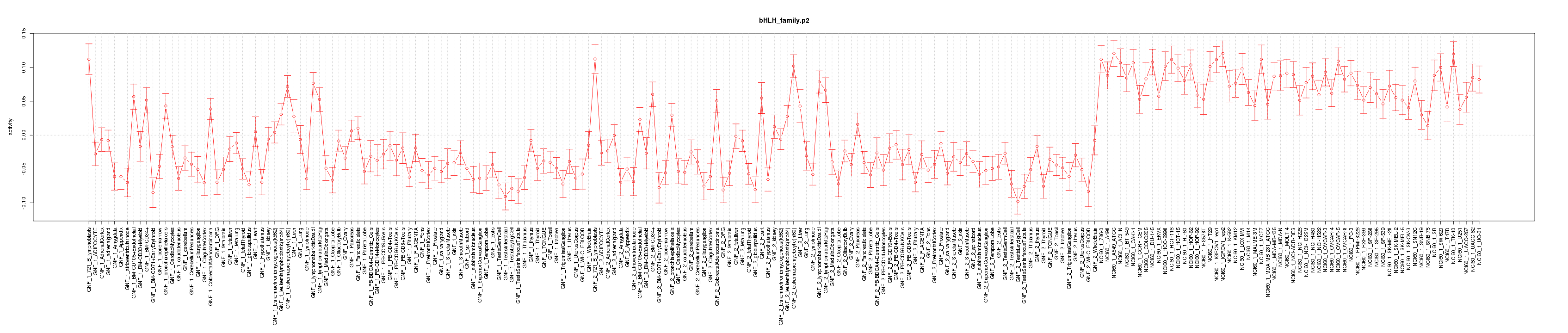
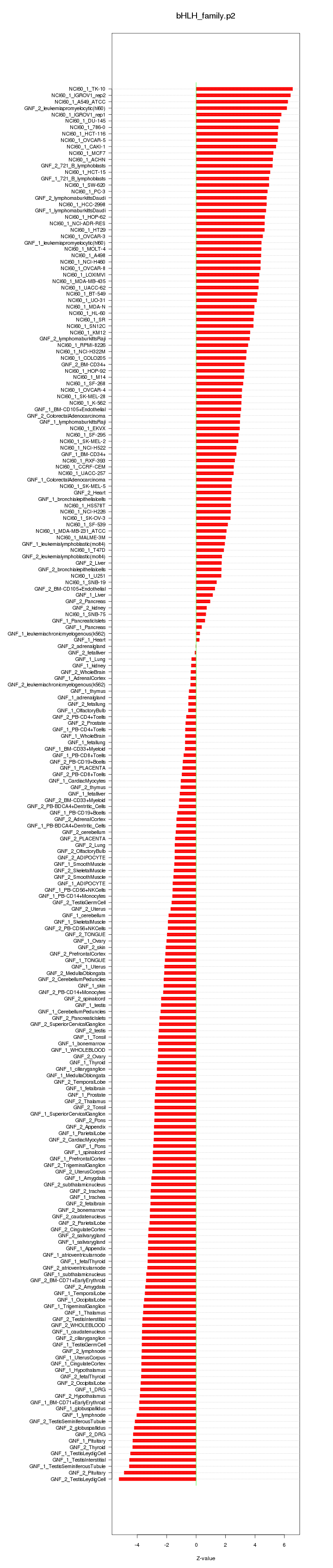
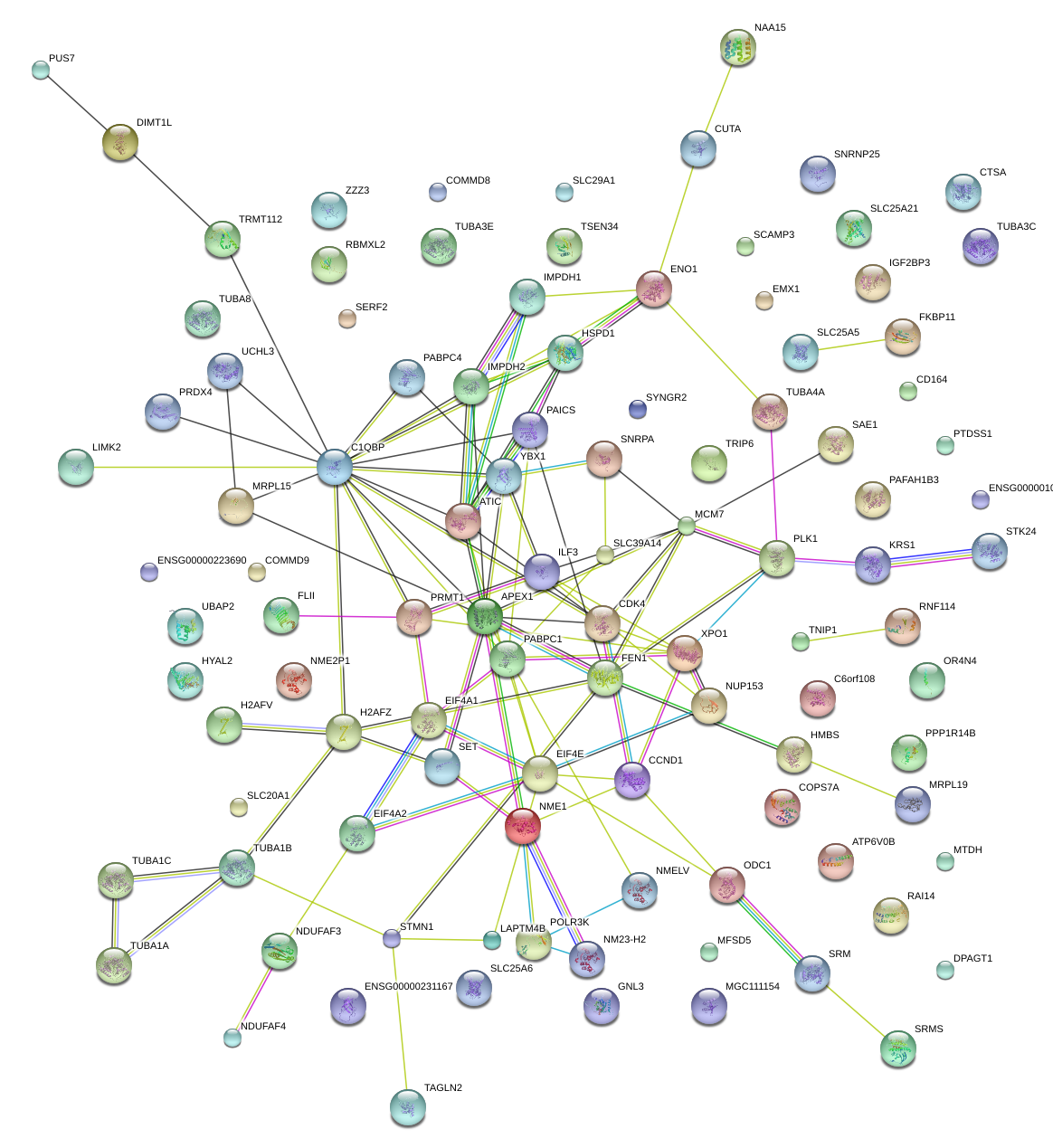
|
1.25
|
3.85e-51
|
GO:0009987
|
cellular process
|
|
1.46
|
1.71e-48
|
GO:0044237
|
cellular metabolic process
|
|
1.38
|
6.40e-41
|
GO:0008152
|
metabolic process
|
|
1.40
|
2.65e-36
|
GO:0044238
|
primary metabolic process
|
|
1.63
|
1.12e-34
|
GO:0006807
|
nitrogen compound metabolic process
|
|
1.64
|
4.99e-34
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
|
1.50
|
2.11e-33
|
GO:0044260
|
cellular macromolecule metabolic process
|
|
1.66
|
7.04e-31
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
|
1.42
|
1.12e-27
|
GO:0043170
|
macromolecule metabolic process
|
|
1.68
|
1.03e-24
|
GO:0090304
|
nucleic acid metabolic process
|
|
1.68
|
1.13e-19
|
GO:0010467
|
gene expression
|
|
1.70
|
3.51e-19
|
GO:0016070
|
RNA metabolic process
|
|
1.55
|
1.14e-17
|
GO:0044249
|
cellular biosynthetic process
|
|
1.52
|
1.04e-16
|
GO:0009058
|
biosynthetic process
|
|
1.41
|
1.22e-15
|
GO:0071840
|
cellular component organization or biogenesis
|
|
1.47
|
7.00e-15
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
|
2.08
|
1.13e-14
|
GO:0006396
|
RNA processing
|
|
1.73
|
1.75e-13
|
GO:0044085
|
cellular component biogenesis
|
|
1.37
|
4.32e-12
|
GO:0016043
|
cellular component organization
|
|
1.45
|
5.56e-12
|
GO:0044267
|
cellular protein metabolic process
|
|
1.54
|
6.60e-12
|
GO:0044281
|
small molecule metabolic process
|
|
1.41
|
1.32e-10
|
GO:0071842
|
cellular component organization at cellular level
|
|
1.54
|
1.98e-10
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
|
1.52
|
4.87e-10
|
GO:0009059
|
macromolecule biosynthetic process
|
|
1.98
|
1.33e-09
|
GO:0016071
|
mRNA metabolic process
|
|
1.61
|
1.23e-08
|
GO:0044248
|
cellular catabolic process
|
|
1.53
|
1.71e-08
|
GO:0009056
|
catabolic process
|
|
2.58
|
2.25e-08
|
GO:0022613
|
ribonucleoprotein complex biogenesis
|
|
3.01
|
3.28e-08
|
GO:0042254
|
ribosome biogenesis
|
|
2.49
|
7.10e-08
|
GO:0071843
|
cellular component biogenesis at cellular level
|
|
1.48
|
8.33e-08
|
GO:0006996
|
organelle organization
|
|
1.40
|
1.21e-07
|
GO:0048523
|
negative regulation of cellular process
|
|
1.33
|
1.21e-07
|
GO:0019538
|
protein metabolic process
|
|
1.60
|
2.79e-07
|
GO:0022607
|
cellular component assembly
|
|
2.22
|
5.40e-07
|
GO:0034660
|
ncRNA metabolic process
|
|
2.05
|
6.06e-07
|
GO:0044419
|
interspecies interaction between organisms
|
|
2.47
|
8.07e-07
|
GO:0034470
|
ncRNA processing
|
|
1.68
|
1.04e-06
|
GO:0046907
|
intracellular transport
|
|
2.12
|
1.09e-06
|
GO:0006412
|
translation
|
|
1.77
|
1.33e-06
|
GO:0009057
|
macromolecule catabolic process
|
|
1.35
|
1.73e-06
|
GO:0048522
|
positive regulation of cellular process
|
|
1.28
|
1.74e-06
|
GO:0051179
|
localization
|
|
1.36
|
1.95e-06
|
GO:0048519
|
negative regulation of biological process
|
|
1.15
|
3.36e-06
|
GO:0065007
|
biological regulation
|
|
1.63
|
3.38e-06
|
GO:0043933
|
macromolecular complex subunit organization
|
|
1.63
|
3.93e-06
|
GO:0033554
|
cellular response to stress
|
|
1.81
|
4.77e-06
|
GO:0044265
|
cellular macromolecule catabolic process
|
|
1.94
|
7.52e-06
|
GO:0006397
|
mRNA processing
|
|
1.55
|
7.70e-06
|
GO:0007049
|
cell cycle
|
|
1.65
|
9.97e-06
|
GO:0071844
|
cellular component assembly at cellular level
|
|
1.48
|
1.20e-05
|
GO:0051641
|
cellular localization
|
|
1.73
|
1.45e-05
|
GO:0071822
|
protein complex subunit organization
|
|
1.31
|
1.79e-05
|
GO:0048518
|
positive regulation of biological process
|
|
1.15
|
2.28e-05
|
GO:0050789
|
regulation of biological process
|
|
1.72
|
2.59e-05
|
GO:0006259
|
DNA metabolic process
|
|
1.16
|
3.05e-05
|
GO:0050794
|
regulation of cellular process
|
|
1.51
|
3.22e-05
|
GO:0051649
|
establishment of localization in cell
|
|
1.73
|
3.98e-05
|
GO:0055086
|
nucleobase, nucleoside and nucleotide metabolic process
|
|
1.54
|
4.13e-05
|
GO:0009892
|
negative regulation of metabolic process
|
|
1.28
|
4.23e-05
|
GO:0006810
|
transport
|
|
1.56
|
4.95e-05
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
|
2.00
|
6.46e-05
|
GO:0008380
|
RNA splicing
|
|
1.28
|
6.53e-05
|
GO:0051234
|
establishment of localization
|
|
1.65
|
9.65e-05
|
GO:0046483
|
heterocycle metabolic process
|
|
1.56
|
1.19e-04
|
GO:0008219
|
cell death
|
|
2.50
|
1.20e-04
|
GO:0007005
|
mitochondrion organization
|
|
2.57
|
1.25e-04
|
GO:0006403
|
RNA localization
|
|
2.89
|
1.30e-04
|
GO:0006364
|
rRNA processing
|
|
1.75
|
1.33e-04
|
GO:0034621
|
cellular macromolecular complex subunit organization
|
|
2.58
|
1.61e-04
|
GO:0050657
|
nucleic acid transport
|
|
2.58
|
1.61e-04
|
GO:0050658
|
RNA transport
|
|
2.58
|
1.61e-04
|
GO:0051236
|
establishment of RNA localization
|
|
1.55
|
1.65e-04
|
GO:0016265
|
death
|
|
2.19
|
1.82e-04
|
GO:0000375
|
RNA splicing, via transesterification reactions
|
|
1.43
|
2.33e-04
|
GO:0033036
|
macromolecule localization
|
|
1.79
|
2.64e-04
|
GO:0072521
|
purine-containing compound metabolic process
|
|
2.41
|
3.63e-04
|
GO:0015931
|
nucleobase, nucleoside, nucleotide and nucleic acid transport
|
|
1.52
|
3.74e-04
|
GO:0031324
|
negative regulation of cellular metabolic process
|
|
2.73
|
5.32e-04
|
GO:0016072
|
rRNA metabolic process
|
|
1.59
|
5.85e-04
|
GO:0065003
|
macromolecular complex assembly
|
|
1.82
|
6.90e-04
|
GO:0044271
|
cellular nitrogen compound biosynthetic process
|
|
1.43
|
9.32e-04
|
GO:0031325
|
positive regulation of cellular metabolic process
|
|
2.16
|
9.42e-04
|
GO:0022411
|
cellular component disassembly
|
|
2.16
|
9.42e-04
|
GO:0071845
|
cellular component disassembly at cellular level
|
|
1.30
|
1.12e-03
|
GO:0042221
|
response to chemical stimulus
|
|
2.04
|
1.26e-03
|
GO:0051186
|
cofactor metabolic process
|
|
2.12
|
1.41e-03
|
GO:0000377
|
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile
|
|
2.12
|
1.41e-03
|
GO:0000398
|
nuclear mRNA splicing, via spliceosome
|
|
1.45
|
1.63e-03
|
GO:0008104
|
protein localization
|
|
1.60
|
1.88e-03
|
GO:0051726
|
regulation of cell cycle
|
|
1.87
|
2.32e-03
|
GO:0051325
|
interphase
|
|
1.71
|
2.40e-03
|
GO:0008283
|
cell proliferation
|
|
1.87
|
2.67e-03
|
GO:0051329
|
interphase of mitotic cell cycle
|
|
1.65
|
3.42e-03
|
GO:0006753
|
nucleoside phosphate metabolic process
|
|
1.65
|
3.42e-03
|
GO:0009117
|
nucleotide metabolic process
|
|
1.81
|
3.58e-03
|
GO:0009259
|
ribonucleotide metabolic process
|
|
1.69
|
3.80e-03
|
GO:0016044
|
cellular membrane organization
|
|
1.69
|
4.66e-03
|
GO:0061024
|
membrane organization
|
|
1.72
|
4.77e-03
|
GO:0016032
|
viral reproduction
|
|
4.09
|
4.78e-03
|
GO:0043488
|
regulation of mRNA stability
|
|
1.27
|
4.88e-03
|
GO:0006950
|
response to stress
|
|
2.75
|
4.93e-03
|
GO:0006839
|
mitochondrial transport
|
|
1.39
|
5.00e-03
|
GO:0009893
|
positive regulation of metabolic process
|
|
1.48
|
5.56e-03
|
GO:0045184
|
establishment of protein localization
|
|
1.49
|
5.84e-03
|
GO:0022402
|
cell cycle process
|
|
1.19
|
7.14e-03
|
GO:0051716
|
cellular response to stimulus
|
|
1.93
|
7.25e-03
|
GO:0032269
|
negative regulation of cellular protein metabolic process
|
|
1.86
|
7.99e-03
|
GO:0010608
|
posttranscriptional regulation of gene expression
|
|
3.95
|
7.99e-03
|
GO:0043487
|
regulation of RNA stability
|
|
1.44
|
8.14e-03
|
GO:0032774
|
RNA biosynthetic process
|
|
1.42
|
8.24e-03
|
GO:0042981
|
regulation of apoptosis
|
|
1.47
|
9.08e-03
|
GO:0015031
|
protein transport
|
|
1.53
|
1.00e-02
|
GO:0022403
|
cell cycle phase
|
|
1.44
|
1.07e-02
|
GO:0042127
|
regulation of cell proliferation
|
|
1.92
|
1.09e-02
|
GO:0018130
|
heterocycle biosynthetic process
|
|
1.89
|
1.24e-02
|
GO:0051248
|
negative regulation of protein metabolic process
|
|
1.41
|
1.36e-02
|
GO:0043067
|
regulation of programmed cell death
|
|
1.41
|
1.42e-02
|
GO:0010941
|
regulation of cell death
|
|
2.07
|
1.45e-02
|
GO:0006732
|
coenzyme metabolic process
|
|
1.62
|
1.67e-02
|
GO:0006461
|
protein complex assembly
|
|
1.59
|
1.68e-02
|
GO:0044282
|
small molecule catabolic process
|
|
1.77
|
1.84e-02
|
GO:0009150
|
purine ribonucleotide metabolic process
|
|
1.74
|
1.85e-02
|
GO:0006520
|
cellular amino acid metabolic process
|
|
3.39
|
1.86e-02
|
GO:0006413
|
translational initiation
|
|
2.92
|
1.94e-02
|
GO:0009119
|
ribonucleoside metabolic process
|
|
1.29
|
1.95e-02
|
GO:0043412
|
macromolecule modification
|
|
2.42
|
2.02e-02
|
GO:0006414
|
translational elongation
|
|
1.61
|
2.20e-02
|
GO:0070271
|
protein complex biogenesis
|
|
1.23
|
2.32e-02
|
GO:0048731
|
system development
|
|
1.66
|
2.40e-02
|
GO:0044106
|
cellular amine metabolic process
|
|
1.59
|
2.70e-02
|
GO:0044283
|
small molecule biosynthetic process
|
|
1.58
|
2.85e-02
|
GO:0006974
|
response to DNA damage stimulus
|
|
1.67
|
2.93e-02
|
GO:0006163
|
purine nucleotide metabolic process
|
|
1.59
|
2.95e-02
|
GO:0009308
|
amine metabolic process
|
|
1.21
|
3.01e-02
|
GO:0048856
|
anatomical structure development
|
|
1.55
|
3.09e-02
|
GO:0000278
|
mitotic cell cycle
|
|
5.64
|
4.15e-02
|
GO:0042451
|
purine nucleoside biosynthetic process
|
|
5.64
|
4.15e-02
|
GO:0042455
|
ribonucleoside biosynthetic process
|
|
5.64
|
4.15e-02
|
GO:0046129
|
purine ribonucleoside biosynthetic process
|
|
1.47
|
4.19e-02
|
GO:0016192
|
vesicle-mediated transport
|
|
2.29
|
4.30e-02
|
GO:0032984
|
macromolecular complex disassembly
|
|
2.29
|
4.30e-02
|
GO:0034623
|
cellular macromolecular complex disassembly
|
|
1.61
|
4.31e-02
|
GO:0044262
|
cellular carbohydrate metabolic process
|
|
3.91
|
4.91e-02
|
GO:0009156
|
ribonucleoside monophosphate biosynthetic process
|