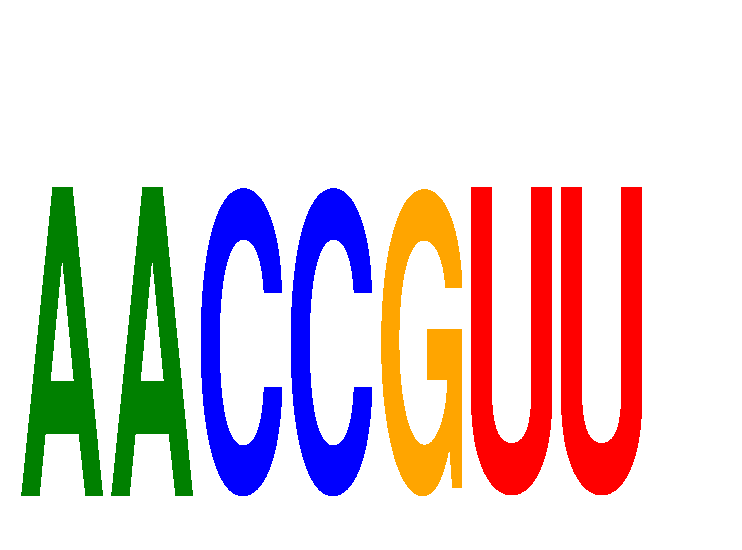Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AACCGUU
Z-value: 0.03
miRNA associated with seed AACCGUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-451a
|
MIMAT0001631 |
Activity profile of AACCGUU motif
Sorted Z-values of AACCGUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_231577532 | 0.11 |
ENST00000258418.5
|
CAB39
|
calcium binding protein 39 |
| chr19_-_10679644 | 0.08 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr8_-_101965146 | 0.05 |
ENST00000395957.2
ENST00000395948.2 ENST00000457309.1 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr16_+_8891670 | 0.02 |
ENST00000268261.4
ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2
|
phosphomannomutase 2 |
| chr17_+_30771279 | 0.02 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr9_-_135819987 | 0.01 |
ENST00000298552.3
ENST00000403810.1 |
TSC1
|
tuberous sclerosis 1 |
| chr11_-_8680383 | 0.01 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |