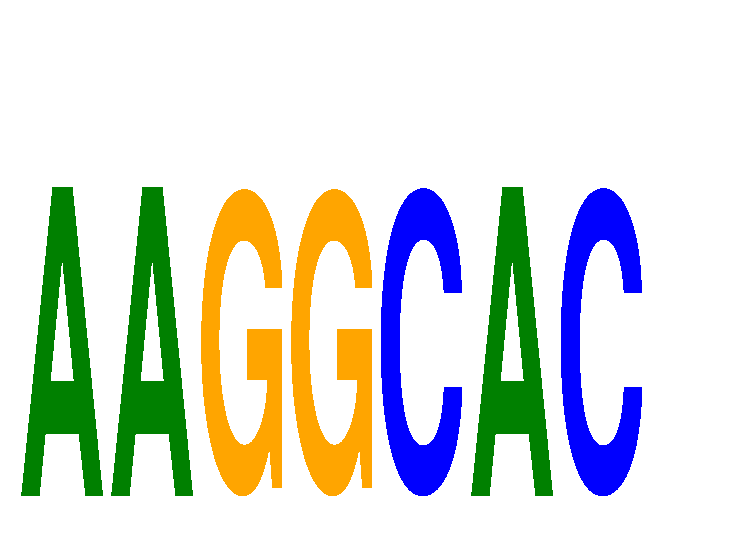Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AAGGCAC
Z-value: 1.79
miRNA associated with seed AAGGCAC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-124-3p.1
|
MIMAT0000422 |
Activity profile of AAGGCAC motif
Sorted Z-values of AAGGCAC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_33246722 | 22.95 |
ENST00000437302.1
ENST00000396033.2 |
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr10_+_17271266 | 22.55 |
ENST00000224237.5
|
VIM
|
vimentin |
| chr2_-_161350305 | 21.55 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr1_+_182992545 | 17.40 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr1_+_223900034 | 17.39 |
ENST00000295006.5
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr21_-_44846999 | 16.64 |
ENST00000270162.6
|
SIK1
|
salt-inducible kinase 1 |
| chr22_-_50746027 | 16.45 |
ENST00000425954.1
ENST00000449103.1 |
PLXNB2
|
plexin B2 |
| chr2_-_64881018 | 16.30 |
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2 |
| chr3_-_120170052 | 15.81 |
ENST00000295633.3
|
FSTL1
|
follistatin-like 1 |
| chr2_+_109204909 | 15.68 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr6_+_138725343 | 15.22 |
ENST00000607197.1
ENST00000367697.3 |
HEBP2
|
heme binding protein 2 |
| chr1_-_154943212 | 15.20 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr4_-_122618095 | 14.87 |
ENST00000515017.1
ENST00000501272.2 ENST00000296511.5 |
ANXA5
|
annexin A5 |
| chr7_+_116165754 | 14.79 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr19_-_50143452 | 13.49 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr7_+_98972298 | 13.47 |
ENST00000252725.5
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa |
| chr9_-_110251836 | 13.18 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr22_-_36784035 | 13.10 |
ENST00000216181.5
|
MYH9
|
myosin, heavy chain 9, non-muscle |
| chr12_-_58146048 | 12.97 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr6_-_36515177 | 12.86 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr7_+_17338239 | 12.85 |
ENST00000242057.4
|
AHR
|
aryl hydrocarbon receptor |
| chr15_-_101792137 | 12.78 |
ENST00000254190.3
|
CHSY1
|
chondroitin sulfate synthase 1 |
| chr19_+_39138271 | 12.46 |
ENST00000252699.2
|
ACTN4
|
actinin, alpha 4 |
| chr2_-_37899323 | 12.33 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr12_-_65146636 | 12.05 |
ENST00000418919.2
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chrX_-_109561294 | 11.89 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr8_-_49833978 | 11.83 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr15_-_55562582 | 11.62 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_+_797392 | 11.52 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr6_-_109703663 | 11.50 |
ENST00000368961.5
|
CD164
|
CD164 molecule, sialomucin |
| chr11_+_60681346 | 11.02 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr5_+_65440032 | 10.79 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr10_-_30024716 | 10.34 |
ENST00000375398.2
ENST00000375400.3 |
SVIL
|
supervillin |
| chr5_+_149340282 | 10.26 |
ENST00000286298.4
|
SLC26A2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr5_+_82767284 | 10.23 |
ENST00000265077.3
|
VCAN
|
versican |
| chr16_-_84651673 | 10.19 |
ENST00000262428.4
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chr7_+_77166592 | 10.17 |
ENST00000248594.6
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr14_+_70078303 | 10.17 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247 |
| chr12_-_15942309 | 10.04 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr7_-_6523755 | 10.01 |
ENST00000436575.1
ENST00000258739.4 |
DAGLB
KDELR2
|
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr11_+_832944 | 10.01 |
ENST00000322008.4
ENST00000397421.1 ENST00000529810.1 ENST00000526693.1 ENST00000525333.1 ENST00000524748.1 ENST00000527341.1 |
CD151
|
CD151 molecule (Raph blood group) |
| chr16_+_69221028 | 9.98 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr8_+_98656336 | 9.96 |
ENST00000336273.3
|
MTDH
|
metadherin |
| chr2_+_69969106 | 9.96 |
ENST00000409920.1
ENST00000394295.4 ENST00000536030.1 |
ANXA4
|
annexin A4 |
| chr1_+_165796753 | 9.93 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr7_+_66386204 | 9.93 |
ENST00000341567.4
ENST00000607045.1 |
TMEM248
|
transmembrane protein 248 |
| chr7_+_142960505 | 9.91 |
ENST00000409500.3
ENST00000443571.2 ENST00000358406.5 ENST00000479303.1 |
GSTK1
|
glutathione S-transferase kappa 1 |
| chr3_+_69812877 | 9.71 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr15_+_90931450 | 9.50 |
ENST00000268182.5
ENST00000560738.1 ENST00000560418.1 |
IQGAP1
|
IQ motif containing GTPase activating protein 1 |
| chr10_-_74856608 | 9.28 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr17_-_7297833 | 9.24 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr5_-_131563501 | 9.22 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr1_+_110091189 | 9.20 |
ENST00000369851.4
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr4_+_129730779 | 9.16 |
ENST00000226319.6
|
PHF17
|
jade family PHD finger 1 |
| chr2_+_219264466 | 9.14 |
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr16_-_85045131 | 8.88 |
ENST00000313732.4
|
ZDHHC7
|
zinc finger, DHHC-type containing 7 |
| chr9_-_124132483 | 8.85 |
ENST00000286713.2
ENST00000538954.1 ENST00000347359.2 |
STOM
|
stomatin |
| chr18_+_19749386 | 8.82 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr17_+_7123125 | 8.80 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr2_-_153574480 | 8.76 |
ENST00000410080.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr17_-_62658186 | 8.73 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr5_-_98262240 | 8.71 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr2_-_43453734 | 8.71 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr12_+_96588143 | 8.67 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr17_-_40540377 | 8.60 |
ENST00000404395.3
ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr6_-_134639180 | 8.56 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr8_-_37756972 | 8.38 |
ENST00000330843.4
ENST00000522727.1 ENST00000287263.4 |
RAB11FIP1
|
RAB11 family interacting protein 1 (class I) |
| chr17_-_41174424 | 8.33 |
ENST00000355653.3
|
VAT1
|
vesicle amine transport 1 |
| chr3_-_182698381 | 8.31 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr15_+_45315302 | 8.29 |
ENST00000267814.9
|
SORD
|
sorbitol dehydrogenase |
| chr5_+_172410757 | 8.23 |
ENST00000519374.1
ENST00000519911.1 ENST00000265093.4 ENST00000517669.1 |
ATP6V0E1
|
ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 |
| chr2_+_28615669 | 8.22 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr10_+_89419370 | 8.20 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr7_+_106809406 | 8.17 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr20_+_361261 | 8.15 |
ENST00000217233.3
|
TRIB3
|
tribbles pseudokinase 3 |
| chr5_-_39425068 | 8.14 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr6_-_4135825 | 8.12 |
ENST00000380118.3
ENST00000413766.2 ENST00000361538.2 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr11_-_22851367 | 8.08 |
ENST00000354193.4
|
SVIP
|
small VCP/p97-interacting protein |
| chrX_+_49028265 | 8.02 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr10_-_3827417 | 8.01 |
ENST00000497571.1
ENST00000542957.1 |
KLF6
|
Kruppel-like factor 6 |
| chrX_+_123095155 | 7.96 |
ENST00000371160.1
ENST00000435103.1 |
STAG2
|
stromal antigen 2 |
| chr6_+_7107999 | 7.90 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr6_-_86352642 | 7.90 |
ENST00000355238.6
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr6_+_143929307 | 7.88 |
ENST00000427704.2
ENST00000305766.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr1_-_225840747 | 7.82 |
ENST00000366843.2
ENST00000366844.3 |
ENAH
|
enabled homolog (Drosophila) |
| chr3_+_37903432 | 7.72 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr10_-_81965307 | 7.63 |
ENST00000537102.1
ENST00000372231.3 ENST00000438331.1 ENST00000422982.3 ENST00000360615.4 ENST00000265447.4 |
ANXA11
|
annexin A11 |
| chr6_+_16129308 | 7.59 |
ENST00000356840.3
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chr5_+_75699040 | 7.57 |
ENST00000274364.6
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr20_-_43977055 | 7.50 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chrX_+_41192595 | 7.47 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr1_-_150947343 | 7.44 |
ENST00000271688.6
ENST00000368954.5 |
CERS2
|
ceramide synthase 2 |
| chr2_+_88991162 | 7.42 |
ENST00000283646.4
|
RPIA
|
ribose 5-phosphate isomerase A |
| chr1_-_38325256 | 7.39 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr16_+_67063036 | 7.30 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr20_-_10654639 | 7.29 |
ENST00000254958.5
|
JAG1
|
jagged 1 |
| chr6_+_37137939 | 7.27 |
ENST00000373509.5
|
PIM1
|
pim-1 oncogene |
| chr6_-_13814663 | 7.24 |
ENST00000359495.2
ENST00000379170.4 |
MCUR1
|
mitochondrial calcium uniporter regulator 1 |
| chr18_+_29077990 | 7.23 |
ENST00000261590.8
|
DSG2
|
desmoglein 2 |
| chr3_-_133969437 | 7.18 |
ENST00000460933.1
ENST00000296084.4 |
RYK
|
receptor-like tyrosine kinase |
| chr8_+_98881268 | 7.17 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr16_-_11680791 | 7.14 |
ENST00000571976.1
ENST00000413364.2 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr6_+_56954867 | 7.02 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chr2_+_178257372 | 7.00 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr5_+_151151471 | 6.99 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr14_+_103058948 | 6.95 |
ENST00000262241.6
|
RCOR1
|
REST corepressor 1 |
| chr1_+_64058939 | 6.89 |
ENST00000371084.3
|
PGM1
|
phosphoglucomutase 1 |
| chr1_-_222885770 | 6.87 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr12_-_58240470 | 6.77 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chrX_-_77150985 | 6.76 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr10_-_33623564 | 6.75 |
ENST00000374875.1
ENST00000374822.4 |
NRP1
|
neuropilin 1 |
| chr17_+_28705921 | 6.75 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr19_-_45908292 | 6.73 |
ENST00000360957.5
ENST00000592134.1 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr7_-_100860851 | 6.69 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr20_+_49126881 | 6.69 |
ENST00000371621.3
ENST00000541713.1 |
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr1_+_193091080 | 6.69 |
ENST00000367435.3
|
CDC73
|
cell division cycle 73 |
| chr3_-_72496035 | 6.69 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chr14_+_21538429 | 6.61 |
ENST00000298694.4
ENST00000555038.1 |
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chrX_+_131157290 | 6.59 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr1_+_40723779 | 6.55 |
ENST00000372759.3
|
ZMPSTE24
|
zinc metallopeptidase STE24 |
| chr6_-_41909561 | 6.54 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr3_-_160283348 | 6.51 |
ENST00000334256.4
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3) |
| chr9_+_137218362 | 6.51 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr21_+_35445827 | 6.47 |
ENST00000608209.1
ENST00000381151.3 |
SLC5A3
SLC5A3
|
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr10_-_27443155 | 6.36 |
ENST00000427324.1
ENST00000326799.3 |
YME1L1
|
YME1-like 1 ATPase |
| chr1_+_26856236 | 6.33 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr21_-_46293586 | 6.31 |
ENST00000445724.2
ENST00000397887.3 |
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein |
| chr7_+_39663061 | 6.31 |
ENST00000005257.2
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr19_-_42759300 | 6.30 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr2_+_46769798 | 6.30 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr9_+_123970052 | 6.27 |
ENST00000373823.3
|
GSN
|
gelsolin |
| chr9_+_114423615 | 6.26 |
ENST00000374293.4
|
GNG10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr12_+_111843749 | 6.24 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr7_-_105162652 | 6.24 |
ENST00000356362.2
ENST00000469408.1 |
PUS7
|
pseudouridylate synthase 7 homolog (S. cerevisiae) |
| chr20_+_42086525 | 6.22 |
ENST00000244020.3
|
SRSF6
|
serine/arginine-rich splicing factor 6 |
| chr8_+_110346546 | 6.22 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chrX_-_10645773 | 6.20 |
ENST00000453318.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr1_+_36348790 | 6.17 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr6_-_52441713 | 6.17 |
ENST00000182527.3
|
TRAM2
|
translocation associated membrane protein 2 |
| chr10_+_72575643 | 6.10 |
ENST00000373202.3
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr3_+_152879985 | 6.08 |
ENST00000323534.2
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chr5_+_122110691 | 6.05 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr7_-_27183263 | 6.00 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr12_+_104359576 | 5.99 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr6_+_35995488 | 5.98 |
ENST00000229795.3
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr2_-_26467557 | 5.91 |
ENST00000380649.3
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr12_+_121124599 | 5.81 |
ENST00000228506.3
|
MLEC
|
malectin |
| chr19_-_33793430 | 5.77 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr3_+_5229356 | 5.76 |
ENST00000256497.4
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr1_+_32645269 | 5.76 |
ENST00000373610.3
|
TXLNA
|
taxilin alpha |
| chr22_-_26986045 | 5.74 |
ENST00000442495.1
ENST00000440953.1 ENST00000450022.1 ENST00000338754.4 |
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr19_-_10697895 | 5.70 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr1_+_112162381 | 5.69 |
ENST00000433097.1
ENST00000369709.3 ENST00000436150.2 |
RAP1A
|
RAP1A, member of RAS oncogene family |
| chr21_-_34852304 | 5.69 |
ENST00000542230.2
|
TMEM50B
|
transmembrane protein 50B |
| chr1_+_7831323 | 5.63 |
ENST00000054666.6
|
VAMP3
|
vesicle-associated membrane protein 3 |
| chr17_+_57784826 | 5.58 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr1_+_173446405 | 5.56 |
ENST00000340385.5
|
PRDX6
|
peroxiredoxin 6 |
| chr1_-_115259337 | 5.54 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr8_+_56014949 | 5.53 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr4_-_54930790 | 5.50 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr16_+_447209 | 5.49 |
ENST00000382940.4
ENST00000219479.2 |
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr6_-_79787902 | 5.49 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr10_-_75173785 | 5.45 |
ENST00000535178.1
ENST00000372921.5 ENST00000372919.4 |
ANXA7
|
annexin A7 |
| chr1_+_116184566 | 5.41 |
ENST00000355485.2
ENST00000369510.4 |
VANGL1
|
VANGL planar cell polarity protein 1 |
| chrX_+_9983602 | 5.41 |
ENST00000380861.4
|
WWC3
|
WWC family member 3 |
| chr5_-_131826457 | 5.37 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr11_+_64948665 | 5.35 |
ENST00000533820.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr20_+_48552908 | 5.34 |
ENST00000244061.2
|
RNF114
|
ring finger protein 114 |
| chr11_+_125462690 | 5.34 |
ENST00000392708.4
ENST00000529196.1 ENST00000531491.1 |
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr5_-_137878887 | 5.33 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr1_+_47799446 | 5.32 |
ENST00000371873.5
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr3_+_187930719 | 5.26 |
ENST00000312675.4
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr20_+_61569463 | 5.23 |
ENST00000266069.3
|
GID8
|
GID complex subunit 8 |
| chr5_-_133968529 | 5.21 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr7_-_137686791 | 5.20 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr8_+_95732095 | 5.18 |
ENST00000414645.2
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr6_+_12012536 | 5.18 |
ENST00000379388.2
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr18_-_18691739 | 5.11 |
ENST00000399799.2
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr11_+_76494253 | 5.07 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr22_-_50946113 | 5.04 |
ENST00000216080.5
ENST00000474879.2 ENST00000380796.3 |
LMF2
|
lipase maturation factor 2 |
| chr19_+_10828724 | 5.04 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr1_-_236228403 | 5.03 |
ENST00000366595.3
|
NID1
|
nidogen 1 |
| chr22_+_21996549 | 5.03 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr11_+_20385327 | 4.99 |
ENST00000451739.2
ENST00000532505.1 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr1_-_16482554 | 4.98 |
ENST00000358432.5
|
EPHA2
|
EPH receptor A2 |
| chr2_-_152684977 | 4.94 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr10_+_72164135 | 4.90 |
ENST00000373218.4
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr10_+_88516396 | 4.86 |
ENST00000372037.3
|
BMPR1A
|
bone morphogenetic protein receptor, type IA |
| chr3_+_180630090 | 4.81 |
ENST00000357559.4
ENST00000305586.7 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr6_-_53213780 | 4.73 |
ENST00000304434.6
ENST00000370918.4 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr2_-_97405775 | 4.71 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr13_-_110959478 | 4.69 |
ENST00000543140.1
ENST00000375820.4 |
COL4A1
|
collagen, type IV, alpha 1 |
| chr1_+_224544552 | 4.66 |
ENST00000465271.1
ENST00000366858.3 |
CNIH4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr5_+_109025067 | 4.66 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr12_+_1800179 | 4.65 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr1_-_95392635 | 4.64 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr11_-_87908600 | 4.61 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chr2_-_26101374 | 4.61 |
ENST00000435504.4
|
ASXL2
|
additional sex combs like 2 (Drosophila) |
| chr2_-_174830430 | 4.60 |
ENST00000310015.6
ENST00000455789.2 |
SP3
|
Sp3 transcription factor |
| chr18_-_19284724 | 4.58 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGCAC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 5.0 | GO:0051604 | protein maturation(GO:0051604) |
| 4.9 | 14.8 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 4.6 | 23.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) regulation of collagen catabolic process(GO:0010710) |
| 4.4 | 13.2 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 4.4 | 13.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 4.2 | 16.6 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 4.2 | 12.5 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 4.1 | 12.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 4.0 | 12.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 3.9 | 11.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 3.7 | 18.5 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 3.7 | 11.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 3.1 | 3.1 | GO:0060926 | cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 3.1 | 21.7 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 2.8 | 2.8 | GO:0035826 | hypotonic response(GO:0006971) rubidium ion transport(GO:0035826) cellular hypotonic response(GO:0071476) regulation of rubidium ion transport(GO:2000680) negative regulation of rubidium ion transport(GO:2000681) regulation of rubidium ion transmembrane transporter activity(GO:2000686) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 2.8 | 2.8 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 2.8 | 8.3 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 2.7 | 8.0 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 2.6 | 5.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 2.5 | 10.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 2.5 | 9.9 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 2.5 | 7.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 2.4 | 4.9 | GO:0048382 | mesendoderm development(GO:0048382) |
| 2.4 | 7.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 2.4 | 14.3 | GO:2000644 | regulation of receptor catabolic process(GO:2000644) |
| 2.3 | 7.0 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 2.2 | 6.7 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 2.2 | 8.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 2.2 | 15.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 2.2 | 6.5 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 2.2 | 8.6 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 2.1 | 8.5 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 2.1 | 8.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 2.0 | 8.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 2.0 | 8.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 2.0 | 13.8 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 2.0 | 7.9 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 1.9 | 11.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.9 | 5.7 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 1.9 | 5.7 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 1.9 | 7.5 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 1.9 | 20.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 1.8 | 7.2 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 1.7 | 5.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.7 | 1.7 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 1.7 | 5.1 | GO:0003383 | apical constriction(GO:0003383) |
| 1.7 | 5.0 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 1.5 | 3.1 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 1.5 | 7.5 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 1.5 | 6.0 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 1.5 | 4.5 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.5 | 7.4 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) negative regulation of glial cell migration(GO:1903976) |
| 1.5 | 7.4 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.4 | 7.2 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 1.4 | 5.8 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 1.4 | 10.0 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 1.4 | 6.8 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 1.3 | 8.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 1.3 | 5.4 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 1.3 | 4.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 1.3 | 3.9 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 1.3 | 6.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 1.3 | 3.8 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 1.3 | 6.3 | GO:1903923 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) protein processing in phagocytic vesicle(GO:1900756) regulation of plasma membrane raft polarization(GO:1903906) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 1.2 | 18.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 1.2 | 2.4 | GO:1901656 | glycoside transport(GO:1901656) |
| 1.2 | 8.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.2 | 6.0 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 1.2 | 3.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 1.2 | 3.5 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 1.2 | 9.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 1.1 | 7.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 1.1 | 13.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) |
| 1.1 | 10.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 1.1 | 3.3 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 1.1 | 31.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.0 | 6.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 1.0 | 4.0 | GO:0016240 | autophagosome docking(GO:0016240) |
| 1.0 | 6.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 1.0 | 5.9 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.9 | 12.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.9 | 2.8 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.9 | 2.8 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.9 | 3.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.9 | 7.3 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.9 | 1.8 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.9 | 5.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.9 | 6.3 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.9 | 1.8 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.9 | 8.9 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.9 | 1.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.9 | 6.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.9 | 8.8 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.9 | 12.3 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.9 | 7.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.9 | 7.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.9 | 17.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.9 | 4.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.8 | 1.7 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.8 | 14.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.8 | 2.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.8 | 3.3 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.8 | 0.8 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.8 | 2.5 | GO:2000276 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.8 | 2.5 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.8 | 7.4 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.8 | 3.3 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.8 | 4.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.8 | 3.1 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.8 | 1.6 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.8 | 5.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.8 | 4.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.8 | 4.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.7 | 2.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.7 | 2.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.7 | 5.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.7 | 5.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.7 | 8.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.7 | 2.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.7 | 5.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.7 | 3.5 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.7 | 6.9 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.7 | 2.1 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.7 | 2.7 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.7 | 6.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.7 | 6.8 | GO:0015791 | polyol transport(GO:0015791) |
| 0.7 | 2.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.7 | 4.0 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.6 | 3.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.6 | 0.6 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.6 | 19.7 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.6 | 6.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.6 | 13.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.6 | 1.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.6 | 6.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.6 | 1.8 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.6 | 6.7 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.6 | 1.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.6 | 3.0 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.6 | 3.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.6 | 7.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.6 | 3.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.6 | 3.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.6 | 9.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.6 | 4.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.6 | 5.8 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.6 | 2.3 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.6 | 20.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.5 | 3.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.5 | 1.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.5 | 10.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.5 | 20.1 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.5 | 22.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.5 | 1.6 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.5 | 7.6 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.5 | 1.5 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.5 | 5.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 3.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.5 | 6.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.5 | 6.8 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.5 | 18.2 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.5 | 2.9 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.5 | 1.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.5 | 3.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.5 | 0.9 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.5 | 3.3 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.5 | 5.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.5 | 6.4 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.4 | 5.8 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.4 | 1.8 | GO:0006109 | regulation of carbohydrate metabolic process(GO:0006109) |
| 0.4 | 8.0 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.4 | 1.8 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.4 | 13.2 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.4 | 1.3 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.4 | 3.4 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.4 | 3.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.4 | 1.3 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.4 | 5.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.4 | 2.5 | GO:0051643 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) secretory granule localization(GO:0032252) endoplasmic reticulum localization(GO:0051643) |
| 0.4 | 3.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.4 | 7.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.4 | 11.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.4 | 2.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 2.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.4 | 2.8 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.4 | 10.3 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.4 | 7.9 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.4 | 2.0 | GO:0051208 | sequestering of calcium ion(GO:0051208) release of sequestered calcium ion into cytosol(GO:0051209) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.4 | 2.3 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.4 | 1.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.4 | 3.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 2.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 4.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.4 | 5.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.4 | 5.3 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.4 | 1.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.4 | 3.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.4 | 3.7 | GO:0071383 | steroid hormone mediated signaling pathway(GO:0043401) cellular response to steroid hormone stimulus(GO:0071383) |
| 0.4 | 1.8 | GO:1902109 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.4 | 6.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.4 | 6.7 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.3 | 4.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 1.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.3 | 5.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 3.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 1.4 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.3 | 1.3 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.3 | 2.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 1.7 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.3 | 2.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.3 | 1.3 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.3 | 5.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.3 | 2.8 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.3 | 18.2 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.3 | 2.8 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.3 | 8.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 1.8 | GO:0030421 | defecation(GO:0030421) |
| 0.3 | 1.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 | 4.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.3 | 3.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.5 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.3 | 3.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.3 | 2.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 11.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.3 | 13.0 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.3 | 9.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 5.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 1.4 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.3 | 1.4 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 3.0 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.3 | 6.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.3 | 8.8 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.3 | 4.4 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.3 | 7.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.3 | 0.5 | GO:0060978 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.3 | 2.4 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.3 | 0.8 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.3 | 10.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.3 | 1.6 | GO:0070489 | T cell activation(GO:0042110) leukocyte aggregation(GO:0070486) T cell aggregation(GO:0070489) lymphocyte aggregation(GO:0071593) |
| 0.3 | 4.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 4.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 7.9 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.2 | 5.4 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.2 | 1.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.2 | 2.0 | GO:0043086 | negative regulation of catalytic activity(GO:0043086) |
| 0.2 | 1.0 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 10.5 | GO:2000142 | regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.2 | 9.8 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 2.3 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 0.9 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 10.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 4.0 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 3.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 12.9 | GO:0030888 | regulation of B cell proliferation(GO:0030888) |
| 0.2 | 4.0 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.2 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 3.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 6.0 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.2 | 2.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.8 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 0.6 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 3.9 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.2 | 4.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 2.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 1.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 2.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 1.0 | GO:0072176 | nephric duct development(GO:0072176) nephric duct morphogenesis(GO:0072178) |
| 0.2 | 21.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 1.9 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.2 | 1.3 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 1.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 2.0 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.2 | 0.5 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 0.9 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.2 | 2.0 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 1.9 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 1.4 | GO:0046349 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 2.3 | GO:2000637 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 1.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 2.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 1.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.2 | 8.0 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.2 | 7.7 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.2 | 1.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 2.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.2 | 0.9 | GO:0090175 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.2 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.6 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.6 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.7 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 1.2 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 2.0 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 4.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 5.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 3.2 | GO:0006605 | protein targeting(GO:0006605) |
| 0.1 | 4.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 2.0 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 1.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.5 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.1 | 3.1 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.1 | 4.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 1.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 1.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.6 | GO:0050790 | regulation of catalytic activity(GO:0050790) |
| 0.1 | 1.2 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) regulation of RNA export from nucleus(GO:0046831) |
| 0.1 | 1.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 4.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 1.0 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 2.8 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 4.0 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 4.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 3.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 1.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 3.7 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.1 | 1.4 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 4.9 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 1.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 3.9 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.1 | 6.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 4.0 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 2.4 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 2.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.9 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.2 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 | 3.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 1.5 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.1 | 1.7 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.9 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.1 | 0.1 | GO:0003157 | endocardium development(GO:0003157) |
| 0.1 | 8.9 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.5 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.2 | GO:0035963 | cellular response to interleukin-13(GO:0035963) |
| 0.1 | 2.1 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 1.2 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.1 | 3.4 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.1 | 0.1 | GO:1903410 | nitric oxide production involved in inflammatory response(GO:0002537) lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.1 | 1.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 3.9 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 4.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.6 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.1 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 3.7 | GO:0038111 | interleukin-7-mediated signaling pathway(GO:0038111) response to interleukin-7(GO:0098760) cellular response to interleukin-7(GO:0098761) |
| 0.1 | 7.8 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 8.7 | GO:0042594 | response to starvation(GO:0042594) |
| 0.1 | 2.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.0 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 1.3 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.3 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 1.2 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 1.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.8 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 2.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0060973 | pericardium morphogenesis(GO:0003344) cell migration involved in heart development(GO:0060973) |
| 0.0 | 2.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.7 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.4 | GO:1902001 | fatty acid transmembrane transport(GO:1902001) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.3 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.0 | 3.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.3 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 4.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 2.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 5.1 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:2000146 | negative regulation of cell motility(GO:2000146) |
| 0.0 | 0.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.8 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 1.1 | GO:0001649 | osteoblast differentiation(GO:0001649) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.0 | GO:0034680 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha3-beta1 complex(GO:0034667) integrin alpha8-beta1 complex(GO:0034678) integrin alpha10-beta1 complex(GO:0034680) |
| 5.0 | 14.9 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 4.4 | 13.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 4.4 | 17.4 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 3.2 | 12.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 3.1 | 9.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 2.5 | 15.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 2.4 | 14.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.6 | 9.9 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.6 | 4.7 | GO:0097447 | dendritic tree(GO:0097447) |
| 1.5 | 4.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.4 | 10.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.3 | 9.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.2 | 8.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.2 | 13.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.2 | 34.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.2 | 7.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 1.2 | 9.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.2 | 4.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.2 | 3.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.2 | 8.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 1.1 | 15.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.1 | 7.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.1 | 5.5 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 1.1 | 3.3 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 1.1 | 13.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.0 | 12.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.9 | 4.6 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.8 | 7.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.8 | 2.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.8 | 3.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.7 | 2.9 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.7 | 13.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.7 | 8.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.7 | 10.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 6.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.6 | 3.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.6 | 8.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.6 | 2.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.6 | 4.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.6 | 3.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.6 | 2.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.5 | 5.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.5 | 3.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.5 | 5.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.5 | 11.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.5 | 3.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.5 | 1.9 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.5 | 6.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 2.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 5.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 16.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 4.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.4 | 8.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 1.3 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.4 | 1.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.4 | 17.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.4 | 1.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.4 | 6.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.4 | 3.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.4 | 2.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.4 | 10.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.4 | 8.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.4 | 3.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.4 | 59.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.3 | 35.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.3 | 2.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 2.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 3.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 1.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 6.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 13.0 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 14.9 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 1.3 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.3 | 0.9 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.3 | 1.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 9.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 8.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 3.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 5.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 4.3 | GO:0001741 | XY body(GO:0001741) |
| 0.3 | 19.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 2.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 7.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 3.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 2.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.3 | 2.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 0.8 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.2 | 3.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 21.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 4.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 1.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 12.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 1.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 6.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.2 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 8.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 11.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 7.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 3.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 2.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 0.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 29.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 11.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.6 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.2 | 5.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 5.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 1.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 6.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 13.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 1.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 1.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 6.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 9.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 28.1 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.2 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 6.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.2 | 80.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.2 | 5.0 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.2 | 0.8 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 5.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 1.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 6.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 22.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 3.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 18.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 11.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 37.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.1 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 5.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 10.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 4.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 26.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 2.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 5.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.5 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.1 | 1.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 32.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 2.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 2.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 3.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 16.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 2.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.4 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.4 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 1.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 4.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 13.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.8 | GO:0070161 | anchoring junction(GO:0070161) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 47.9 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.8 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 4.6 | 23.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 4.3 | 12.8 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 4.0 | 24.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 3.7 | 18.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 3.4 | 10.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 3.2 | 12.9 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 3.2 | 15.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 3.0 | 12.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 2.7 | 8.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 2.6 | 13.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 2.5 | 10.0 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 2.2 | 8.8 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 2.2 | 15.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 2.2 | 15.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 2.1 | 6.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 2.1 | 10.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 2.1 | 6.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 2.0 | 8.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 2.0 | 6.0 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 2.0 | 9.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.9 | 5.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 1.7 | 10.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.5 | 4.5 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.5 | 5.9 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 1.4 | 4.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 1.4 | 24.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 1.4 | 20.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 1.3 | 13.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.3 | 7.8 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 1.2 | 7.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.2 | 12.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.2 | 6.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.2 | 4.6 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 1.1 | 15.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 1.1 | 5.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 1.1 | 3.2 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 1.1 | 4.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 1.1 | 4.3 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
| 1.1 | 14.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.0 | 3.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.0 | 3.1 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 1.0 | 6.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.0 | 3.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 1.0 | 24.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 1.0 | 17.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.9 | 7.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.9 | 2.8 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.9 | 3.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.9 | 4.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.9 | 5.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.9 | 3.5 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.8 | 5.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.8 | 5.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.8 | 3.3 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.8 | 15.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.8 | 4.9 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.8 | 6.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.8 | 7.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.8 | 10.4 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.8 | 6.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.8 | 4.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.8 | 3.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.8 | 6.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.8 | 7.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.7 | 16.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.7 | 2.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.7 | 13.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.7 | 7.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.7 | 2.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.7 | 3.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.7 | 3.3 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.7 | 3.3 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.6 | 6.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.6 | 23.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.6 | 5.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.6 | 5.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 1.8 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.6 | 9.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.6 | 3.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.6 | 4.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.6 | 10.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.6 | 5.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.5 | 6.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.5 | 1.6 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.5 | 2.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 5.6 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.5 | 9.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.5 | 13.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.5 | 6.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 3.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.5 | 8.2 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.5 | 1.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.5 | 5.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 2.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.5 | 2.8 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.5 | 9.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.4 | 4.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.4 | 5.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 1.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 12.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.4 | 2.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.4 | 1.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 6.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 1.7 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.4 | 1.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 13.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.4 | 11.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.4 | 5.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.4 | 0.8 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.4 | 3.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.4 | 2.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 13.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.4 | 15.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.4 | 5.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.4 | 1.8 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.4 | 3.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.4 | 4.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.4 | 5.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.4 | 10.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.3 | 2.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 4.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 15.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 6.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.3 | 1.7 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.3 | 1.0 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 2.9 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.3 | 31.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.3 | 5.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 2.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 4.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 4.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 8.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 3.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 8.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.3 | 1.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.3 | 12.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 4.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 10.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 12.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 1.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.3 | 17.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.3 | 25.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 6.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.3 | 2.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 4.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 4.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 7.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 1.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 3.9 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 4.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 3.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.7 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 22.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 0.9 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.2 | 2.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 2.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 3.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.2 | 7.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.2 | 0.6 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 2.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 2.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 4.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 8.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 3.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 2.6 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.2 | 2.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 11.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 1.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 3.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 2.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 7.1 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.2 | 1.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 7.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 1.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 2.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 1.7 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 0.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 6.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 3.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 3.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 2.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 1.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 3.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 17.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.2 | 3.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.8 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 3.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.9 | GO:0004871 | signal transducer activity(GO:0004871) |
| 0.1 | 6.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 41.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 3.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 20.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 7.4 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 3.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 6.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 2.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 5.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 24.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 2.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 4.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 1.5 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 2.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.9 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 1.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 18.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.3 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.8 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) |
| 0.1 | 3.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 13.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 3.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.1 | 6.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.0 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 2.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 8.3 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.1 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 6.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 8.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 5.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 3.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.9 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 7.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.6 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.6 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 3.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.7 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 2.9 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 50.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.8 | 24.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.8 | 17.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.6 | 38.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.6 | 5.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.6 | 30.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.6 | 24.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.6 | 44.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.6 | 34.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.6 | 22.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.5 | 13.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 7.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 7.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 5.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 22.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 25.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 20.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.4 | 26.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 3.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.3 | 6.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.3 | 12.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 10.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 56.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 11.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 23.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 9.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 3.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 12.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 20.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 7.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 6.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 5.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 6.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 8.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 3.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 2.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 2.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 6.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 3.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 6.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 0.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 11.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 1.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 9.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 5.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 11.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 8.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 7.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 26.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 7.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 3.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 8.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 3.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 38.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 1.2 | 24.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 1.0 | 32.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.9 | 22.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.9 | 24.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.9 | 15.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.8 | 33.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.8 | 7.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.8 | 13.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.8 | 23.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.7 | 13.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.7 | 7.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.6 | 14.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.6 | 22.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.6 | 3.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.6 | 16.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.6 | 0.6 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.5 | 13.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.5 | 11.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.5 | 9.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 16.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.5 | 15.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.5 | 4.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 5.0 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.5 | 10.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 10.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.4 | 9.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 13.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 6.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.4 | 4.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 6.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 3.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 7.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.3 | 14.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 6.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 30.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.3 | 12.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.3 | 6.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 4.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 8.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 9.1 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.3 | 9.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 1.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 5.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 0.8 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.2 | 8.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 1.0 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.2 | 2.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 6.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 22.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 9.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 6.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.2 | 3.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 4.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 3.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 4.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 2.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 7.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 1.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 4.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 7.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 5.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 3.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 3.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 11.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 6.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 11.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 20.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 6.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 11.3 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 1.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 11.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 2.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 5.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 4.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 4.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 3.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 1.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 3.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 2.0 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |