Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
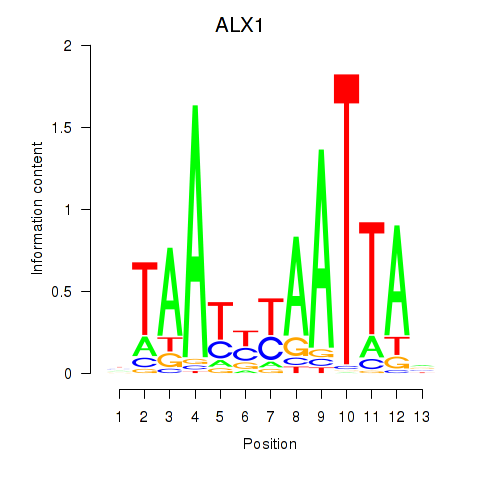
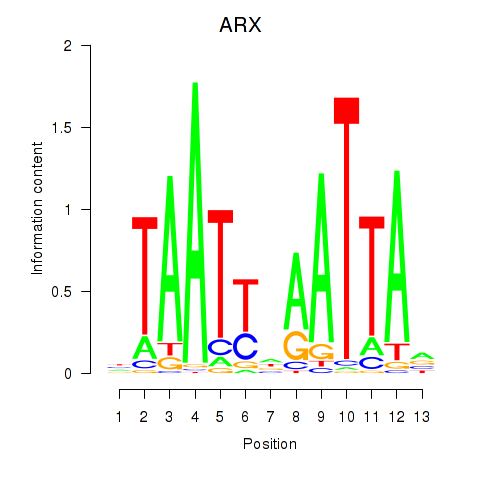
Results for ALX1_ARX
Z-value: 0.38
Transcription factors associated with ALX1_ARX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX1
|
ENSG00000180318.3 | ALX homeobox 1 |
|
ARX
|
ENSG00000004848.6 | aristaless related homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX1 | hg19_v2_chr12_+_85673868_85673885 | 0.05 | 4.3e-01 | Click! |
Activity profile of ALX1_ARX motif
Sorted Z-values of ALX1_ARX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_176733897 | 6.92 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr6_+_34204642 | 5.35 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr1_+_50569575 | 5.06 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_+_102413726 | 4.54 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr12_-_118797475 | 4.34 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chrX_+_119737806 | 3.94 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr1_+_84630645 | 3.78 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr10_-_75226166 | 3.58 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr2_-_55496174 | 3.51 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr18_-_21891460 | 3.01 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr8_+_26150628 | 2.71 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr1_+_206557366 | 2.62 |
ENST00000414007.1
ENST00000419187.2 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr1_-_21377383 | 2.49 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr3_+_138340049 | 2.39 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chrX_+_107288197 | 2.36 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr12_+_104337515 | 2.18 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chrX_+_107288239 | 2.11 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_-_33759699 | 2.07 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_+_145826867 | 1.81 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr8_-_102803163 | 1.75 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr15_+_80733570 | 1.68 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr4_+_113568207 | 1.65 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr3_+_130569429 | 1.60 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr3_-_98241760 | 1.57 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr2_-_145278475 | 1.52 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr7_-_93520259 | 1.51 |
ENST00000222543.5
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr12_-_118628350 | 1.43 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr2_+_75873902 | 1.43 |
ENST00000393909.2
ENST00000358788.6 ENST00000409374.1 |
MRPL19
|
mitochondrial ribosomal protein L19 |
| chr6_-_150067696 | 1.43 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr8_+_107738240 | 1.38 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr10_+_18689637 | 1.34 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr3_-_141747439 | 1.33 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_-_99797522 | 1.31 |
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila) |
| chr5_-_134735568 | 1.24 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr15_+_57511609 | 1.23 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr5_+_36152091 | 1.22 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr7_-_93520191 | 1.21 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr6_-_134639180 | 1.20 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_141747950 | 1.18 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_-_100356551 | 1.17 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr3_-_130465604 | 1.12 |
ENST00000356763.3
|
PIK3R4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr4_-_87028478 | 1.11 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr3_+_140660743 | 1.09 |
ENST00000453248.2
|
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr16_+_24549014 | 1.09 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr7_+_90338712 | 1.06 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr1_-_154600421 | 0.98 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr2_-_70529180 | 0.97 |
ENST00000450256.1
ENST00000037869.3 |
FAM136A
|
family with sequence similarity 136, member A |
| chr21_-_43346790 | 0.95 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr3_-_194188956 | 0.95 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr5_-_9630463 | 0.89 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr19_-_54619006 | 0.87 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr15_-_34331243 | 0.85 |
ENST00000306730.3
|
AVEN
|
apoptosis, caspase activation inhibitor |
| chr15_+_44092784 | 0.82 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chr5_+_140227048 | 0.82 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr3_+_138340067 | 0.82 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr5_+_36152163 | 0.81 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr4_-_83769996 | 0.79 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr8_-_91095099 | 0.78 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr12_-_89746173 | 0.77 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr3_+_140660634 | 0.77 |
ENST00000446041.2
ENST00000507429.1 ENST00000324194.6 |
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr8_-_122653630 | 0.73 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr4_-_143227088 | 0.72 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_244214577 | 0.72 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr10_-_112064665 | 0.69 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr9_+_42671887 | 0.62 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr14_-_101036119 | 0.62 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr5_+_140762268 | 0.62 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr12_+_12878829 | 0.61 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr6_-_41039567 | 0.61 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr3_-_47934234 | 0.61 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr2_-_145275228 | 0.59 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_166930131 | 0.59 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr1_+_158978768 | 0.59 |
ENST00000447473.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_-_94147385 | 0.58 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr2_+_54683419 | 0.58 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr2_+_58655461 | 0.57 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr4_-_174320687 | 0.56 |
ENST00000296506.3
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr13_-_47471155 | 0.51 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr7_-_14880892 | 0.48 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr3_-_164914640 | 0.47 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr3_-_9291063 | 0.43 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr2_+_196313239 | 0.42 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr7_-_82792215 | 0.41 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr1_+_158323755 | 0.36 |
ENST00000368157.1
ENST00000368156.1 ENST00000368155.3 ENST00000368154.1 ENST00000368160.3 ENST00000368161.3 |
CD1E
|
CD1e molecule |
| chr4_+_169013666 | 0.35 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr8_-_61880248 | 0.34 |
ENST00000525556.1
|
AC022182.3
|
AC022182.3 |
| chr1_+_92632542 | 0.33 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr2_+_185463093 | 0.31 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr6_-_111804905 | 0.31 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr14_-_78083112 | 0.30 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr2_+_44001172 | 0.29 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr5_+_137203557 | 0.28 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr7_+_107224364 | 0.27 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr4_-_83765613 | 0.27 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr17_+_68071458 | 0.27 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chrX_+_149887090 | 0.27 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr1_+_104615595 | 0.26 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr8_-_62602327 | 0.26 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr17_+_68071389 | 0.25 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_-_141747459 | 0.23 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr19_-_6393465 | 0.23 |
ENST00000394456.5
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa |
| chr2_+_161993412 | 0.21 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr9_+_135854091 | 0.20 |
ENST00000450530.1
ENST00000534944.1 |
GFI1B
|
growth factor independent 1B transcription repressor |
| chr18_-_19994830 | 0.20 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr8_+_38677850 | 0.19 |
ENST00000518809.1
ENST00000520611.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr3_+_155860751 | 0.18 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr9_-_27005686 | 0.17 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr11_+_95523621 | 0.16 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr12_-_67197760 | 0.15 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr5_+_140743859 | 0.15 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr3_-_124774802 | 0.14 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr11_+_120973375 | 0.14 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr4_-_100356291 | 0.14 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr9_+_77230499 | 0.13 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr9_-_21202204 | 0.13 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr16_+_1728257 | 0.12 |
ENST00000248098.3
ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L
|
hematological and neurological expressed 1-like |
| chr17_-_3030875 | 0.12 |
ENST00000328890.2
|
OR1G1
|
olfactory receptor, family 1, subfamily G, member 1 |
| chr11_-_102709441 | 0.11 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr4_-_143226979 | 0.11 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr3_-_157824292 | 0.10 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr1_-_110933611 | 0.10 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr3_+_39424828 | 0.09 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr12_-_43833515 | 0.08 |
ENST00000549670.1
ENST00000395541.2 |
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr1_+_183774240 | 0.07 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr2_+_161993465 | 0.05 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr18_-_52989525 | 0.05 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr3_+_190105909 | 0.04 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr5_-_111312622 | 0.04 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr5_-_55412774 | 0.03 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr2_+_101437487 | 0.03 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr17_+_7155819 | 0.03 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr6_+_26199737 | 0.03 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr1_+_52682052 | 0.03 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr5_+_137203465 | 0.01 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr16_+_68119247 | 0.01 |
ENST00000575270.1
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX1_ARX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 1.3 | 3.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.8 | 3.8 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.6 | 1.9 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.6 | 3.5 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.5 | 2.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.4 | 1.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.2 | GO:1901837 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.3 | 1.7 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.3 | 1.0 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.3 | 3.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.3 | 1.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.6 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.3 | 2.6 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.3 | 2.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 5.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 1.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 4.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.8 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.2 | 6.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 1.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 2.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 0.7 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 2.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 2.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.5 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.8 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 1.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 2.7 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.3 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.1 | 3.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 2.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.6 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.9 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.4 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 1.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 2.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0090270 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.0 | 0.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.1 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.4 | 3.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 1.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 1.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 6.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 2.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 2.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.2 | 1.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 3.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 2.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 2.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 3.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.9 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0043194 | voltage-gated sodium channel complex(GO:0001518) axon initial segment(GO:0043194) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 2.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.6 | 4.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 5.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.4 | 3.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 1.3 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.3 | 3.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 1.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 0.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 3.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 1.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.8 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 2.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 5.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 1.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 4.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 3.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 3.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 6.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 2.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.6 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 4.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 5.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 3.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 1.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 3.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 3.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |