Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ARID5A
Z-value: 0.94
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.11 | AT-rich interaction domain 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg19_v2_chr2_+_97203082_97203159, hg19_v2_chr2_+_97202480_97202499 | 0.08 | 2.6e-01 | Click! |
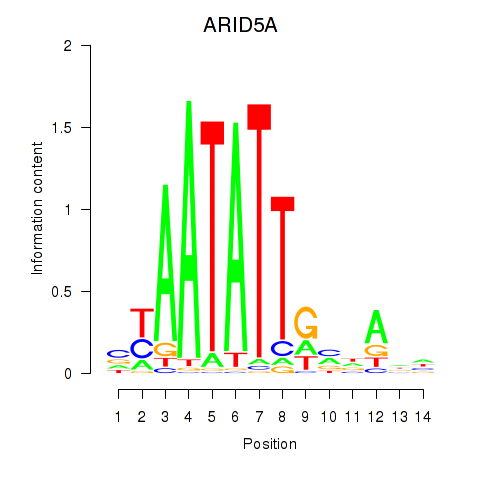
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_88323218 | 21.58 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr3_-_112127981 | 20.54 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr2_-_217724767 | 13.75 |
ENST00000236979.2
|
TNP1
|
transition protein 1 (during histone to protamine replacement) |
| chr3_+_101546827 | 8.71 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr12_+_8995832 | 8.54 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr6_+_150920999 | 8.29 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr16_+_55542910 | 8.17 |
ENST00000262134.5
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2 |
| chr16_-_11375179 | 7.59 |
ENST00000312511.3
|
PRM1
|
protamine 1 |
| chr4_+_106631966 | 6.74 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr2_+_152214098 | 5.44 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr6_-_74161977 | 5.25 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr12_-_91572278 | 5.07 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr19_-_21950362 | 4.77 |
ENST00000358296.6
|
ZNF100
|
zinc finger protein 100 |
| chr1_+_158815588 | 4.62 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr18_+_39766626 | 4.37 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr6_+_151561085 | 4.25 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr10_-_21661870 | 4.15 |
ENST00000433460.1
|
RP11-275N1.1
|
RP11-275N1.1 |
| chr6_-_52705641 | 4.05 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr5_-_131892501 | 3.94 |
ENST00000450655.1
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr10_+_32856764 | 3.91 |
ENST00000375030.2
ENST00000375028.3 |
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr12_-_4758159 | 3.89 |
ENST00000545990.2
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr12_-_8218997 | 3.85 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr2_+_196313239 | 3.76 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr11_+_60197040 | 3.76 |
ENST00000300190.2
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr11_+_60197069 | 3.65 |
ENST00000528905.1
ENST00000528093.1 |
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr2_-_89545079 | 3.63 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr6_+_33048222 | 3.56 |
ENST00000428835.1
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr4_-_71532339 | 3.54 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr11_-_104480019 | 3.51 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr6_+_151561506 | 3.29 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr19_-_57988871 | 3.17 |
ENST00000596831.1
ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9
ZNF772
|
Uncharacterized protein zinc finger protein 772 |
| chrX_+_8432871 | 3.15 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr6_-_33239712 | 3.06 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr5_-_19988339 | 3.04 |
ENST00000382275.1
|
CDH18
|
cadherin 18, type 2 |
| chr13_-_52703187 | 2.84 |
ENST00000355568.4
|
NEK5
|
NIMA-related kinase 5 |
| chr2_-_208989225 | 2.78 |
ENST00000264376.4
|
CRYGD
|
crystallin, gamma D |
| chr22_-_38699003 | 2.78 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr3_-_141747950 | 2.74 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_+_54519242 | 2.73 |
ENST00000234827.1
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr11_-_114466477 | 2.67 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr15_+_93749295 | 2.61 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr14_-_90798418 | 2.50 |
ENST00000354366.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr6_-_133084580 | 2.50 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr5_-_86534822 | 2.45 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr16_+_10479906 | 2.43 |
ENST00000562527.1
ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr3_+_113616317 | 2.42 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr3_-_106959424 | 2.41 |
ENST00000607801.1
ENST00000479612.2 ENST00000484698.1 ENST00000477210.2 ENST00000473636.1 |
LINC00882
|
long intergenic non-protein coding RNA 882 |
| chr11_+_128563652 | 2.39 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr22_-_32766972 | 2.39 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr12_-_91546926 | 2.37 |
ENST00000550758.1
|
DCN
|
decorin |
| chr11_+_55029628 | 2.35 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chr11_-_102595681 | 2.33 |
ENST00000236826.3
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr10_+_114710516 | 2.33 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_-_69171708 | 2.33 |
ENST00000420581.2
|
LMOD3
|
leiomodin 3 (fetal) |
| chr2_-_232395169 | 2.29 |
ENST00000305141.4
|
NMUR1
|
neuromedin U receptor 1 |
| chr15_+_69307028 | 2.27 |
ENST00000388866.3
ENST00000530406.2 |
NOX5
|
NADPH oxidase, EF-hand calcium binding domain 5 |
| chr20_+_37590942 | 2.22 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr6_-_49834209 | 2.21 |
ENST00000507853.1
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr11_-_114466471 | 2.19 |
ENST00000424261.2
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr6_-_49712147 | 2.19 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr19_+_55014085 | 2.17 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr6_+_31707725 | 2.16 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr11_-_62477041 | 2.14 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr18_+_61554932 | 2.13 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr20_+_55904815 | 2.13 |
ENST00000371263.3
ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11
|
SPO11 meiotic protein covalently bound to DSB |
| chr6_-_49834240 | 2.12 |
ENST00000335847.4
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr22_-_32767017 | 2.10 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr6_-_33048483 | 2.09 |
ENST00000419277.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr12_+_10460417 | 2.07 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr8_-_86253888 | 2.06 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr11_-_118305921 | 2.03 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr1_-_203320617 | 2.03 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr17_+_35767301 | 2.02 |
ENST00000225396.6
ENST00000417170.1 ENST00000590005.1 ENST00000590957.1 |
TADA2A
|
transcriptional adaptor 2A |
| chrX_+_41583408 | 1.99 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr3_-_87325728 | 1.97 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr14_+_50291993 | 1.96 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr18_-_5419797 | 1.93 |
ENST00000542146.1
ENST00000427684.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr18_-_53069419 | 1.90 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr14_-_25045446 | 1.88 |
ENST00000216336.2
|
CTSG
|
cathepsin G |
| chr7_-_140482926 | 1.88 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr12_+_9980069 | 1.86 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr22_-_23922448 | 1.85 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr1_-_23504176 | 1.85 |
ENST00000302291.4
|
LUZP1
|
leucine zipper protein 1 |
| chr7_-_80551671 | 1.85 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr11_-_62476965 | 1.84 |
ENST00000405837.1
ENST00000531524.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr7_+_116593292 | 1.82 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr6_-_169654139 | 1.81 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr8_+_11141925 | 1.79 |
ENST00000221086.3
|
MTMR9
|
myotubularin related protein 9 |
| chr4_+_100737954 | 1.74 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr19_+_55014013 | 1.73 |
ENST00000301202.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr4_+_71226468 | 1.73 |
ENST00000226460.4
|
SMR3A
|
submaxillary gland androgen regulated protein 3A |
| chr1_-_150669604 | 1.73 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr13_-_41706864 | 1.70 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr1_-_158656488 | 1.70 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chrX_+_37850026 | 1.68 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr15_-_58571445 | 1.67 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr3_+_134204881 | 1.67 |
ENST00000511574.1
ENST00000337090.3 ENST00000383229.3 |
CEP63
|
centrosomal protein 63kDa |
| chr5_+_159848854 | 1.67 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr12_+_32260085 | 1.66 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr1_+_160336851 | 1.66 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr19_-_44174305 | 1.65 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr17_-_37844267 | 1.63 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr1_+_13910194 | 1.61 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr12_+_72080253 | 1.61 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr3_+_171561127 | 1.59 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr2_-_228244013 | 1.59 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr1_-_216978709 | 1.55 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr17_-_2996290 | 1.54 |
ENST00000331459.1
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2 |
| chr1_-_153013588 | 1.54 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr15_-_52404921 | 1.54 |
ENST00000561198.1
ENST00000260442.3 |
BCL2L10
|
BCL2-like 10 (apoptosis facilitator) |
| chr1_-_46642154 | 1.53 |
ENST00000540385.1
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr6_+_83777374 | 1.52 |
ENST00000349129.2
ENST00000237163.5 ENST00000536812.1 |
DOPEY1
|
dopey family member 1 |
| chr1_+_163039143 | 1.51 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr1_+_104159999 | 1.51 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr5_-_131879205 | 1.50 |
ENST00000231454.1
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr9_-_138391692 | 1.50 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr1_+_115572415 | 1.49 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr3_-_112320749 | 1.48 |
ENST00000610103.1
|
RP11-572C15.6
|
RP11-572C15.6 |
| chr8_+_24298531 | 1.47 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr19_-_44174330 | 1.46 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chrX_+_37639302 | 1.46 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr10_-_15902449 | 1.45 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr3_+_135741576 | 1.45 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr11_-_59950622 | 1.44 |
ENST00000323961.3
ENST00000412309.2 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr16_+_31366455 | 1.43 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr21_-_19775973 | 1.43 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr3_-_194072019 | 1.43 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr10_-_61720640 | 1.42 |
ENST00000521074.1
ENST00000444900.1 |
C10orf40
|
chromosome 10 open reading frame 40 |
| chr17_-_6554877 | 1.42 |
ENST00000225728.3
ENST00000575197.1 |
MED31
|
mediator complex subunit 31 |
| chrX_+_107288239 | 1.42 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr7_-_7782204 | 1.41 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr6_-_49712123 | 1.41 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr1_+_158901329 | 1.39 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chrX_-_100641155 | 1.39 |
ENST00000372880.1
ENST00000308731.7 |
BTK
|
Bruton agammaglobulinemia tyrosine kinase |
| chr4_+_88754069 | 1.39 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr20_-_18447667 | 1.38 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr12_-_96390063 | 1.36 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr11_+_76745385 | 1.35 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr22_+_32754139 | 1.34 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr10_+_114710425 | 1.34 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr8_+_24298597 | 1.34 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr16_+_31366536 | 1.34 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr1_+_248020481 | 1.33 |
ENST00000366481.3
|
TRIM58
|
tripartite motif containing 58 |
| chrX_-_45060135 | 1.32 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr10_+_70847852 | 1.30 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chrX_-_19988382 | 1.27 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr4_-_47983519 | 1.27 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr1_+_241695670 | 1.26 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr6_+_26156551 | 1.25 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr10_+_47746929 | 1.25 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr6_+_32605195 | 1.25 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr11_+_73661364 | 1.24 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr12_-_10542617 | 1.24 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr7_-_19748640 | 1.23 |
ENST00000222567.5
|
TWISTNB
|
TWIST neighbor |
| chr2_+_171034646 | 1.23 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr9_+_116207007 | 1.23 |
ENST00000374140.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr4_+_56212270 | 1.23 |
ENST00000264228.4
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr22_-_29457832 | 1.23 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr12_+_21207503 | 1.22 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr19_+_36132631 | 1.21 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chrX_+_15767971 | 1.19 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr3_+_134204551 | 1.19 |
ENST00000332047.5
ENST00000354446.3 |
CEP63
|
centrosomal protein 63kDa |
| chr1_+_86934526 | 1.18 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr4_+_71248795 | 1.18 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr1_+_178310581 | 1.16 |
ENST00000462775.1
|
RASAL2
|
RAS protein activator like 2 |
| chr5_+_159848807 | 1.15 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr11_-_59950519 | 1.15 |
ENST00000528851.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr18_+_61575200 | 1.14 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr11_-_89653576 | 1.09 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr4_+_68424434 | 1.09 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr5_-_39274617 | 1.09 |
ENST00000510188.1
|
FYB
|
FYN binding protein |
| chr21_+_40824003 | 1.08 |
ENST00000452550.1
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr14_+_39703112 | 1.07 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr15_+_59063478 | 1.07 |
ENST00000559228.1
ENST00000450403.2 |
FAM63B
|
family with sequence similarity 63, member B |
| chr1_+_117963209 | 1.06 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr6_-_33239612 | 1.06 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr1_-_146054494 | 1.06 |
ENST00000401009.2
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr1_+_241695424 | 1.05 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr12_-_15114603 | 1.04 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr22_-_23922410 | 1.03 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr1_+_170904612 | 1.01 |
ENST00000367759.4
ENST00000367758.3 |
MROH9
|
maestro heat-like repeat family member 9 |
| chr2_+_157330081 | 0.99 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr5_-_55412774 | 0.99 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr17_-_2966901 | 0.99 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor, family 1, subfamily D, member 5 |
| chr1_+_104293028 | 0.99 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr20_+_30795664 | 0.98 |
ENST00000375749.3
ENST00000375730.3 ENST00000539210.1 |
POFUT1
|
protein O-fucosyltransferase 1 |
| chr6_-_133055896 | 0.98 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr11_-_112034803 | 0.98 |
ENST00000528832.1
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr12_-_96390108 | 0.96 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chrX_+_70521584 | 0.96 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr10_-_102027420 | 0.96 |
ENST00000354105.4
|
CWF19L1
|
CWF19-like 1, cell cycle control (S. pombe) |
| chr10_+_96443378 | 0.95 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr3_+_148447887 | 0.94 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr7_+_134528635 | 0.94 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr12_+_75874984 | 0.93 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr20_-_18477862 | 0.93 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr4_+_71296204 | 0.93 |
ENST00000413702.1
|
MUC7
|
mucin 7, secreted |
| chr2_-_183106641 | 0.93 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_59950486 | 0.92 |
ENST00000426738.2
ENST00000533023.1 ENST00000420732.2 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr6_+_20534672 | 0.91 |
ENST00000274695.4
ENST00000378624.4 |
CDKAL1
|
CDK5 regulatory subunit associated protein 1-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 2.0 | 8.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 1.8 | 5.4 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 1.4 | 4.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.8 | 3.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.8 | 2.3 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.8 | 2.3 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.7 | 2.7 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.7 | 2.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.6 | 7.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.6 | 3.7 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.5 | 1.6 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.5 | 7.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.5 | 1.4 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.5 | 2.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.5 | 1.4 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.5 | 2.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.4 | 1.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.4 | 1.2 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.4 | 1.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.4 | 2.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 1.9 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 1.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.4 | 1.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 3.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.3 | 1.7 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.3 | 2.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.3 | 1.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.3 | 0.9 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.3 | 1.5 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.3 | 0.9 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 | 4.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 1.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.3 | 0.8 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.3 | 1.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.3 | 3.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 5.2 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.2 | 1.0 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.2 | 1.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 2.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 1.9 | GO:0070943 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.2 | 0.4 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 | 0.8 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.4 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.2 | 4.6 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.2 | 1.0 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.2 | 0.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 2.1 | GO:0090220 | meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.2 | 1.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.2 | 5.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.2 | 0.9 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 | 1.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 0.7 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 1.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 4.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 0.7 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 6.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.3 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.6 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 1.8 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.5 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 2.8 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.9 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 3.9 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.6 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.1 | 0.6 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 2.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 2.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 2.8 | GO:0045143 | homologous chromosome segregation(GO:0045143) |
| 0.1 | 1.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.7 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.1 | 0.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 2.5 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 7.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 0.7 | GO:0032074 | regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 2.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 1.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 2.8 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 1.5 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 0.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 3.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 2.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 5.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.2 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 1.4 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 0.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 2.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.2 | GO:0035989 | tendon development(GO:0035989) tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 1.6 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.7 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 2.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 1.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 7.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.8 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 2.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 1.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 5.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) |
| 0.0 | 1.3 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 2.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 1.2 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.4 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.8 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 1.5 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.0 | 1.9 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 0.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 1.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 1.8 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 | 1.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.7 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 1.3 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 6.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.4 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.2 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.8 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 0.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.6 | GO:0030097 | hemopoiesis(GO:0030097) |
| 0.0 | 0.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.9 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.3 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.0 | 0.8 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 15.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 1.0 | 4.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.6 | 3.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.5 | 7.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.5 | 0.9 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.4 | 8.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 3.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 1.7 | GO:0033648 | host cell cytoplasm(GO:0030430) host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) host cell cytoplasm part(GO:0033655) tubulin complex(GO:0045298) |
| 0.3 | 0.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.3 | 3.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 1.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 1.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 1.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 1.2 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 2.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.0 | GO:0042611 | MHC protein complex(GO:0042611) MHC class I protein complex(GO:0042612) |
| 0.1 | 1.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 8.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 8.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 3.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 4.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 8.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 1.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 5.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 27.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 7.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.2 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 1.8 | 5.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.0 | 3.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.7 | 3.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.7 | 2.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.7 | 3.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.6 | 2.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.5 | 7.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 1.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.5 | 1.4 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.4 | 1.7 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.4 | 1.2 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.4 | 2.3 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.4 | 1.5 | GO:0016160 | amylase activity(GO:0016160) |
| 0.3 | 3.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 1.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 3.8 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.3 | 2.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.0 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.2 | 1.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 1.2 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.2 | 4.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 1.9 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.2 | 1.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 5.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 2.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.9 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.2 | 5.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 0.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 3.8 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.7 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 1.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 1.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 3.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 5.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 12.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 7.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 1.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 1.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 8.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 4.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 2.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 2.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.0 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 1.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 2.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 5.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.7 | GO:0001105 | RNA polymerase II activating transcription factor binding(GO:0001102) RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 4.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.4 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 8.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.2 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 3.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.2 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 5.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 4.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 3.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 23.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 4.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 6.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 6.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 8.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.3 | 3.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 7.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 0.9 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.2 | 2.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 2.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 4.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 5.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 6.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 4.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 2.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 8.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 1.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 3.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 2.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 1.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.0 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |