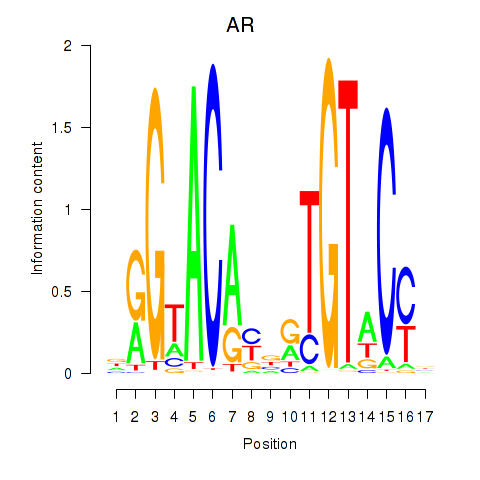
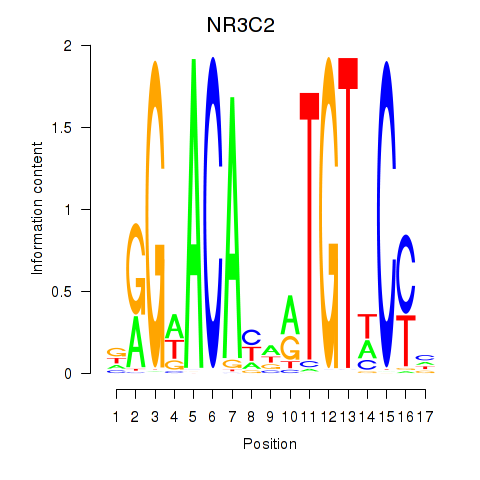
|
chr16_+_56659687
Show fit
|
62.81 |
ENST00000568293.1
ENST00000330439.6
|
MT1E
|
metallothionein 1E
|
|
chr16_+_56716336
Show fit
|
46.85 |
ENST00000394485.4
ENST00000562939.1
|
MT1X
|
metallothionein 1X
|
|
chr3_-_58563094
Show fit
|
37.44 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr16_+_56642041
Show fit
|
35.97 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A
|
|
chr16_+_56642489
Show fit
|
34.57 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A
|
|
chr1_+_22979474
Show fit
|
34.26 |
ENST00000509305.1
|
C1QB
|
complement component 1, q subcomponent, B chain
|
|
chr1_+_22979676
Show fit
|
33.39 |
ENST00000432749.2
ENST00000314933.6
|
C1QB
|
complement component 1, q subcomponent, B chain
|
|
chr12_+_53443680
Show fit
|
32.88 |
ENST00000314250.6
ENST00000451358.1
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr12_+_53443963
Show fit
|
30.26 |
ENST00000546602.1
ENST00000552570.1
ENST00000549700.1
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr19_+_35630926
Show fit
|
29.65 |
ENST00000588081.1
ENST00000589121.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr19_+_35630628
Show fit
|
27.87 |
ENST00000588715.1
ENST00000588607.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr14_-_21493884
Show fit
|
25.04 |
ENST00000556974.1
ENST00000554419.1
ENST00000298687.5
ENST00000397858.1
ENST00000360463.3
ENST00000350792.3
ENST00000397847.2
|
NDRG2
|
NDRG family member 2
|
|
chr14_-_21493649
Show fit
|
21.98 |
ENST00000553442.1
ENST00000555869.1
ENST00000556457.1
ENST00000397844.2
ENST00000554415.1
|
NDRG2
|
NDRG family member 2
|
|
chr16_+_56666563
Show fit
|
19.89 |
ENST00000570233.1
|
MT1M
|
metallothionein 1M
|
|
chr2_+_238395879
Show fit
|
18.73 |
ENST00000445024.2
ENST00000338530.4
ENST00000409373.1
|
MLPH
|
melanophilin
|
|
chr7_-_94285402
Show fit
|
16.76 |
ENST00000428696.2
ENST00000445866.2
|
SGCE
|
sarcoglycan, epsilon
|
|
chr2_+_238395803
Show fit
|
16.20 |
ENST00000264605.3
|
MLPH
|
melanophilin
|
|
chr16_+_56672571
Show fit
|
14.97 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A
|
|
chr17_-_19290117
Show fit
|
14.20 |
ENST00000497081.2
|
MFAP4
|
microfibrillar-associated protein 4
|
|
chr7_-_94285511
Show fit
|
13.92 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon
|
|
chr17_-_19290483
Show fit
|
12.37 |
ENST00000395592.2
ENST00000299610.4
|
MFAP4
|
microfibrillar-associated protein 4
|
|
chr5_+_140186647
Show fit
|
10.64 |
ENST00000512229.2
ENST00000356878.4
ENST00000530339.1
|
PCDHA4
|
protocadherin alpha 4
|
|
chr19_+_35630344
Show fit
|
10.07 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr18_+_56530136
Show fit
|
9.18 |
ENST00000591083.1
|
ZNF532
|
zinc finger protein 532
|
|
chr7_+_94285637
Show fit
|
8.52 |
ENST00000482108.1
ENST00000488574.1
|
PEG10
|
paternally expressed 10
|
|
chrX_-_6453159
Show fit
|
8.27 |
ENST00000381089.3
ENST00000398729.1
|
VCX3A
|
variable charge, X-linked 3A
|
|
chr7_-_94285472
Show fit
|
7.74 |
ENST00000437425.2
ENST00000447873.1
ENST00000415788.2
|
SGCE
|
sarcoglycan, epsilon
|
|
chr5_-_149792295
Show fit
|
7.29 |
ENST00000518797.1
ENST00000524315.1
ENST00000009530.7
ENST00000377795.3
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chrY_-_16098393
Show fit
|
7.00 |
ENST00000250825.4
|
VCY
|
variable charge, Y-linked
|
|
chr11_+_18287801
Show fit
|
6.58 |
ENST00000532858.1
ENST00000405158.2
|
SAA1
|
serum amyloid A1
|
|
chr12_+_51318513
Show fit
|
6.33 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A
|
|
chrX_-_8139308
Show fit
|
6.29 |
ENST00000317103.4
|
VCX2
|
variable charge, X-linked 2
|
|
chr2_-_242576864
Show fit
|
6.21 |
ENST00000407315.1
|
THAP4
|
THAP domain containing 4
|
|
chr18_+_7754957
Show fit
|
6.13 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr11_+_18287721
Show fit
|
6.08 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1
|
|
chrY_+_16168097
Show fit
|
5.51 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B
|
|
chr19_-_8408139
Show fit
|
5.43 |
ENST00000330915.3
ENST00000593649.1
ENST00000595639.1
|
KANK3
|
KN motif and ankyrin repeat domains 3
|
|
chr17_-_61996136
Show fit
|
5.33 |
ENST00000342364.4
|
GH1
|
growth hormone 1
|
|
chr11_-_236326
Show fit
|
4.90 |
ENST00000525237.1
ENST00000532956.1
ENST00000525319.1
ENST00000524564.1
ENST00000382743.4
|
SIRT3
|
sirtuin 3
|
|
chr2_+_102953608
Show fit
|
4.85 |
ENST00000311734.2
ENST00000409584.1
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr12_+_66582919
Show fit
|
4.74 |
ENST00000545837.1
ENST00000457197.2
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr8_-_49834299
Show fit
|
4.73 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2
|
|
chr8_-_49833978
Show fit
|
4.69 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2
|
|
chr15_+_42694573
Show fit
|
4.67 |
ENST00000397200.4
ENST00000569827.1
|
CAPN3
|
calpain 3, (p94)
|
|
chr17_-_61996160
Show fit
|
4.35 |
ENST00000458650.2
ENST00000351388.4
ENST00000323322.5
|
GH1
|
growth hormone 1
|
|
chr12_-_11002063
Show fit
|
4.15 |
ENST00000544994.1
ENST00000228811.4
ENST00000540107.1
|
PRR4
|
proline rich 4 (lacrimal)
|
|
chr3_-_15382875
Show fit
|
4.12 |
ENST00000408919.3
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr14_+_24616588
Show fit
|
4.01 |
ENST00000324103.6
ENST00000559260.1
|
RNF31
|
ring finger protein 31
|
|
chr5_-_179499086
Show fit
|
3.62 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130
|
|
chr16_-_4852915
Show fit
|
3.55 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr16_+_67207838
Show fit
|
3.52 |
ENST00000566871.1
ENST00000268605.7
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr5_-_179499108
Show fit
|
3.50 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130
|
|
chr17_-_61996192
Show fit
|
3.32 |
ENST00000392824.4
|
CSHL1
|
chorionic somatomammotropin hormone-like 1
|
|
chr12_-_10251603
Show fit
|
3.16 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr17_-_61959202
Show fit
|
2.91 |
ENST00000449787.2
ENST00000456543.2
ENST00000423893.2
ENST00000332800.7
|
GH2
|
growth hormone 2
|
|
chr16_-_66584059
Show fit
|
2.89 |
ENST00000417693.3
ENST00000544898.1
ENST00000569718.1
ENST00000527284.1
ENST00000299697.7
ENST00000451102.2
|
TK2
|
thymidine kinase 2, mitochondrial
|
|
chr16_+_67207872
Show fit
|
2.76 |
ENST00000563258.1
ENST00000568146.1
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr6_-_52774464
Show fit
|
2.69 |
ENST00000370968.1
ENST00000211122.3
|
GSTA3
|
glutathione S-transferase alpha 3
|
|
chr19_-_14117074
Show fit
|
2.67 |
ENST00000588885.1
ENST00000254325.4
|
RFX1
|
regulatory factor X, 1 (influences HLA class II expression)
|
|
chr4_-_8442438
Show fit
|
2.63 |
ENST00000356406.5
ENST00000413009.2
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr12_+_125549925
Show fit
|
2.41 |
ENST00000316519.6
|
AACS
|
acetoacetyl-CoA synthetase
|
|
chrX_+_17755563
Show fit
|
2.21 |
ENST00000380045.3
ENST00000380041.3
ENST00000380043.3
ENST00000398080.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila)
|
|
chr5_-_179498703
Show fit
|
2.21 |
ENST00000522208.2
|
RNF130
|
ring finger protein 130
|
|
chrX_+_153029633
Show fit
|
2.18 |
ENST00000538966.1
ENST00000361971.5
ENST00000538776.1
ENST00000538543.1
|
PLXNB3
|
plexin B3
|
|
chr17_-_16472483
Show fit
|
2.04 |
ENST00000395824.1
ENST00000448349.2
ENST00000395825.3
|
ZNF287
|
zinc finger protein 287
|
|
chr6_-_111927062
Show fit
|
1.96 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr11_-_506739
Show fit
|
1.78 |
ENST00000529306.1
ENST00000438658.2
ENST00000527485.1
ENST00000397615.2
ENST00000397614.1
|
RNH1
|
ribonuclease/angiogenin inhibitor 1
|
|
chr7_+_94139105
Show fit
|
1.77 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1
|
|
chr1_+_76262552
Show fit
|
1.71 |
ENST00000263187.3
|
MSH4
|
mutS homolog 4
|
|
chr15_-_28344439
Show fit
|
1.71 |
ENST00000431101.1
ENST00000445578.1
ENST00000353809.5
ENST00000382996.2
ENST00000354638.3
|
OCA2
|
oculocutaneous albinism II
|
|
chrX_+_2609356
Show fit
|
1.54 |
ENST00000381180.3
ENST00000449611.1
|
CD99
|
CD99 molecule
|
|
chr8_-_21669826
Show fit
|
1.44 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chr3_+_169491171
Show fit
|
1.38 |
ENST00000356716.4
|
MYNN
|
myoneurin
|
|
chrX_+_2609207
Show fit
|
1.37 |
ENST00000381192.3
|
CD99
|
CD99 molecule
|
|
chrX_+_8433376
Show fit
|
1.31 |
ENST00000440654.2
ENST00000381029.4
|
VCX3B
|
variable charge, X-linked 3B
|
|
chr17_+_41323204
Show fit
|
1.22 |
ENST00000542611.1
ENST00000590996.1
ENST00000389312.4
ENST00000589872.1
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr12_+_125549973
Show fit
|
1.21 |
ENST00000536752.1
ENST00000261686.6
|
AACS
|
acetoacetyl-CoA synthetase
|
|
chr13_+_113777105
Show fit
|
1.19 |
ENST00000409306.1
ENST00000375551.3
ENST00000375559.3
|
F10
|
coagulation factor X
|
|
chr1_-_27693349
Show fit
|
1.19 |
ENST00000374040.3
ENST00000357582.2
ENST00000493901.1
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6
|
|
chr22_+_18632666
Show fit
|
1.04 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18
|
|
chr19_+_8117881
Show fit
|
1.00 |
ENST00000390669.3
|
CCL25
|
chemokine (C-C motif) ligand 25
|
|
chr22_+_19939026
Show fit
|
0.97 |
ENST00000406520.3
|
COMT
|
catechol-O-methyltransferase
|
|
chr4_+_87928140
Show fit
|
0.94 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr2_+_27435179
Show fit
|
0.89 |
ENST00000606999.1
ENST00000405489.3
|
ATRAID
|
all-trans retinoic acid-induced differentiation factor
|
|
chr3_+_169490606
Show fit
|
0.86 |
ENST00000349841.5
|
MYNN
|
myoneurin
|
|
chr14_-_106692191
Show fit
|
0.66 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21
|
|
chr16_-_67978016
Show fit
|
0.60 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr1_-_241799232
Show fit
|
0.60 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chrX_+_2609317
Show fit
|
0.57 |
ENST00000381187.3
ENST00000381184.1
|
CD99
|
CD99 molecule
|
|
chr1_-_24469602
Show fit
|
0.55 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr3_-_188665428
Show fit
|
0.54 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1
|
|
chr3_+_13521665
Show fit
|
0.54 |
ENST00000295757.3
ENST00000402259.1
ENST00000402271.1
ENST00000446613.2
ENST00000404548.1
ENST00000404040.1
|
HDAC11
|
histone deacetylase 11
|
|
chr19_+_54704610
Show fit
|
0.50 |
ENST00000302907.4
|
RPS9
|
ribosomal protein S9
|
|
chr8_-_62602327
Show fit
|
0.28 |
ENST00000445642.3
ENST00000517847.2
ENST00000389204.4
ENST00000517661.1
ENST00000517903.1
ENST00000522603.1
ENST00000522349.1
ENST00000522835.1
ENST00000541428.1
ENST00000518306.1
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr13_-_26452679
Show fit
|
0.14 |
ENST00000596729.1
|
AL138815.2
|
Uncharacterized protein
|
|
chr12_-_56615693
Show fit
|
0.08 |
ENST00000394013.2
ENST00000345093.4
ENST00000551711.1
ENST00000552656.1
|
RNF41
|
ring finger protein 41
|