Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
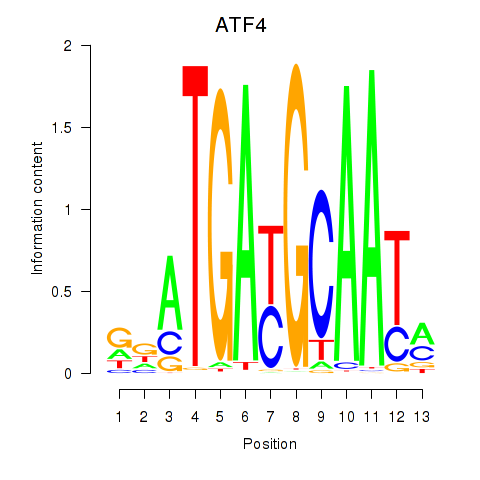
Results for ATF4
Z-value: 1.41
Transcription factors associated with ATF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF4
|
ENSG00000128272.10 | activating transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF4 | hg19_v2_chr22_+_39916558_39916579 | 0.65 | 3.4e-27 | Click! |
Activity profile of ATF4 motif
Sorted Z-values of ATF4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_57624085 | 32.97 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_+_57624119 | 29.04 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr9_-_95055956 | 29.00 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr7_+_30634297 | 26.89 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr5_+_33440802 | 26.63 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
| chr5_+_33441053 | 26.15 |
ENST00000541634.1
ENST00000455217.2 ENST00000414361.2 |
TARS
|
threonyl-tRNA synthetase |
| chr14_-_100841670 | 26.08 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr12_+_57623869 | 25.57 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_-_46662772 | 24.66 |
ENST00000549049.1
ENST00000439706.1 ENST00000398637.5 |
SLC38A1
|
solute carrier family 38, member 1 |
| chr12_+_57623477 | 22.98 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr14_-_100841930 | 20.51 |
ENST00000555031.1
ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr19_-_47287990 | 19.01 |
ENST00000593713.1
ENST00000598022.1 ENST00000434726.2 |
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr17_-_79895097 | 18.53 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr19_-_47288162 | 17.88 |
ENST00000594991.1
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr17_-_79895154 | 16.86 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chrX_+_24072833 | 16.73 |
ENST00000253039.4
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr7_+_99006550 | 16.15 |
ENST00000222969.5
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr5_+_44809027 | 15.98 |
ENST00000507110.1
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr9_+_100745615 | 15.57 |
ENST00000339399.4
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr12_-_92539614 | 14.09 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr7_+_99006232 | 13.56 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr14_+_24563262 | 13.33 |
ENST00000559250.1
ENST00000216780.4 ENST00000560736.1 ENST00000396973.4 ENST00000559837.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr10_+_14880157 | 12.73 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr19_+_33865218 | 12.66 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr7_-_56101826 | 12.49 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chrX_+_77359671 | 11.95 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr9_+_80912059 | 11.91 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chrX_+_77359726 | 11.64 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr14_+_24563510 | 11.23 |
ENST00000545054.2
ENST00000561286.1 ENST00000558096.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr16_-_70323422 | 10.85 |
ENST00000261772.8
|
AARS
|
alanyl-tRNA synthetase |
| chr11_-_3078838 | 10.72 |
ENST00000397111.5
|
CARS
|
cysteinyl-tRNA synthetase |
| chr1_-_220220000 | 10.70 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr11_-_3078616 | 10.19 |
ENST00000401769.3
ENST00000278224.9 ENST00000397114.3 ENST00000380525.4 |
CARS
|
cysteinyl-tRNA synthetase |
| chr15_+_89182178 | 9.86 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr10_+_102106829 | 9.15 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr12_+_57849048 | 9.14 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr11_+_62649158 | 8.99 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr7_-_93633658 | 8.96 |
ENST00000433727.1
|
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr7_-_93633684 | 8.93 |
ENST00000222547.3
ENST00000425626.1 |
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr15_+_89181974 | 8.50 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr17_+_39846114 | 8.42 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chr17_-_77813186 | 8.09 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr11_-_8680383 | 7.62 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr21_-_44495919 | 7.59 |
ENST00000398158.1
|
CBS
|
cystathionine-beta-synthase |
| chr16_+_56970567 | 6.76 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr2_+_46769798 | 6.42 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr1_-_220219775 | 6.27 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr1_+_154244987 | 5.94 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr12_+_50135588 | 5.87 |
ENST00000423828.1
ENST00000550445.1 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_+_77870856 | 5.72 |
ENST00000264893.6
ENST00000502584.1 ENST00000510641.1 |
SEPT11
|
septin 11 |
| chr7_-_99006443 | 5.40 |
ENST00000350498.3
|
PDAP1
|
PDGFA associated protein 1 |
| chr21_-_44495964 | 5.27 |
ENST00000398168.1
ENST00000398165.3 |
CBS
|
cystathionine-beta-synthase |
| chr16_-_18908196 | 4.45 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr2_-_216300784 | 3.67 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr15_+_89182156 | 3.49 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr12_-_25102252 | 3.45 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr17_+_73089382 | 3.41 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chrX_-_133792480 | 3.25 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr17_-_46703826 | 3.23 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr12_-_117318788 | 3.16 |
ENST00000550505.1
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr10_-_101190202 | 3.09 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr15_+_41245160 | 2.94 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr4_-_70725856 | 2.59 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr11_+_118955583 | 2.58 |
ENST00000278715.3
ENST00000536813.1 ENST00000537841.1 ENST00000542729.1 ENST00000546302.1 ENST00000442944.2 ENST00000544387.1 ENST00000543090.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr19_-_49258606 | 1.94 |
ENST00000310160.3
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| chr12_-_25101920 | 1.92 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chrX_+_9431324 | 1.88 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr10_+_63661053 | 1.64 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr3_-_33686743 | 1.44 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr14_-_92413353 | 1.34 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr5_-_145562147 | 1.23 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr1_-_44497024 | 0.97 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr12_-_10324716 | 0.73 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chrX_-_80457385 | 0.61 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr16_+_56965960 | 0.34 |
ENST00000439977.2
ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr3_+_11314099 | 0.33 |
ENST00000446450.2
ENST00000354956.5 ENST00000354449.3 ENST00000419112.1 |
ATG7
|
autophagy related 7 |
| chr1_-_44497118 | 0.28 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr3_-_48130707 | 0.27 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr5_+_174151536 | 0.26 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr22_+_42229100 | 0.14 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr1_+_212782012 | 0.10 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.6 | 52.8 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 15.8 | 110.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 9.7 | 29.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 9.3 | 46.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 9.2 | 36.9 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 7.3 | 21.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 7.0 | 20.9 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 6.7 | 26.9 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 5.9 | 35.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 4.3 | 12.9 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 4.2 | 12.7 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 3.7 | 29.7 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 3.6 | 10.9 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 3.1 | 24.6 | GO:0006116 | NADH oxidation(GO:0006116) |
| 2.7 | 24.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 2.6 | 17.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 1.8 | 9.0 | GO:0060356 | leucine import(GO:0060356) |
| 1.6 | 12.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 1.2 | 3.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.1 | 14.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.0 | 3.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 1.0 | 5.9 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.9 | 25.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.9 | 5.4 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.9 | 2.6 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.8 | 23.6 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.8 | 8.4 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.5 | 15.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.5 | 7.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.4 | 18.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.3 | 5.9 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 6.4 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.2 | 9.2 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.2 | 16.0 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.2 | 2.6 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 3.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 | 2.9 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 3.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.2 | 1.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 0.3 | GO:1901859 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.2 | 1.9 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 9.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.3 | GO:0051795 | positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) BMP signaling pathway involved in heart development(GO:0061312) |
| 0.1 | 8.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 4.2 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 1.6 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.1 | GO:1903121 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 3.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.6 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 1.9 | GO:0016575 | histone deacetylation(GO:0016575) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.8 | 110.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 5.6 | 16.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 2.4 | 17.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.6 | 30.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 1.2 | 7.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.6 | 8.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 21.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 3.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 17.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 7.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 70.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 29.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 23.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 6.4 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 28.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 5.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 46.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 17.9 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 5.7 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 1.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 3.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 71.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 2.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 15.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 10.7 | GO:0043005 | neuron projection(GO:0043005) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.6 | 52.8 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 15.8 | 110.6 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 9.7 | 29.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 9.3 | 46.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 8.8 | 35.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 8.2 | 24.6 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 7.9 | 23.6 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 7.3 | 21.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 7.0 | 20.9 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 5.3 | 36.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 4.3 | 12.9 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 3.6 | 10.9 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 3.1 | 9.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 2.5 | 12.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 2.1 | 6.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 1.7 | 26.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 1.3 | 25.9 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 1.0 | 3.1 | GO:0070546 | phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.0 | 5.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.9 | 17.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.9 | 9.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.9 | 17.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.4 | 1.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) valine-tRNA ligase activity(GO:0004832) |
| 0.4 | 8.1 | GO:0019789 | SUMO transferase activity(GO:0019789) SUMO binding(GO:0032183) |
| 0.4 | 5.9 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.4 | 2.6 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.4 | 12.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 25.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 19.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 15.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.2 | 2.9 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 17.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 9.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 4.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 3.2 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.1 | 16.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 3.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 3.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 2.6 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 3.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 29.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.5 | 23.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 6.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 24.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 9.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 8.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 5.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 205.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 2.2 | 62.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.7 | 70.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.8 | 48.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.7 | 9.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 12.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 21.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 16.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 5.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 6.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 4.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |