Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
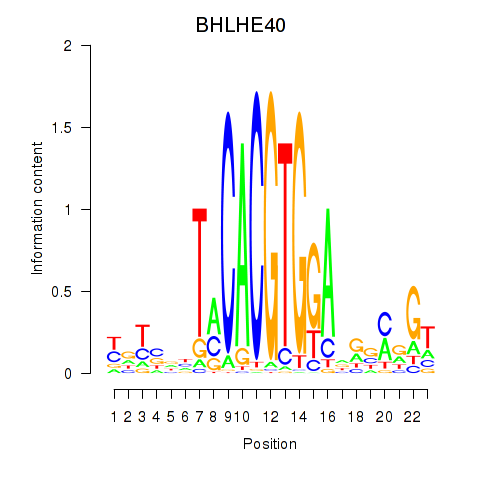
Results for BHLHE40
Z-value: 1.63
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.4 | basic helix-loop-helix family member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE40 | hg19_v2_chr3_+_5020801_5020952 | 0.04 | 5.3e-01 | Click! |
Activity profile of BHLHE40 motif
Sorted Z-values of BHLHE40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_23685653 | 53.23 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr1_+_203830703 | 39.48 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chrX_+_23685563 | 37.35 |
ENST00000379341.4
|
PRDX4
|
peroxiredoxin 4 |
| chr8_+_55047763 | 37.08 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr1_-_43638168 | 32.12 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr2_-_10588630 | 30.69 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr4_+_57301896 | 28.79 |
ENST00000514888.1
ENST00000264221.2 ENST00000505164.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr2_-_10587897 | 28.18 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr19_+_35645618 | 28.12 |
ENST00000392218.2
ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr1_-_43637915 | 27.14 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr4_+_57302297 | 25.93 |
ENST00000399688.3
ENST00000512576.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr19_+_35645817 | 25.14 |
ENST00000423817.3
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr2_-_47143160 | 25.07 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr12_+_66217911 | 24.25 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr12_+_93861264 | 23.37 |
ENST00000549982.1
ENST00000361630.2 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr12_+_93861282 | 23.10 |
ENST00000552217.1
ENST00000393128.4 ENST00000547098.1 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr17_-_61850894 | 22.02 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr2_-_47142884 | 21.41 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr11_+_60609537 | 21.41 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr13_+_50656307 | 20.66 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr6_+_13615554 | 20.53 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chr7_-_107642348 | 19.60 |
ENST00000393561.1
|
LAMB1
|
laminin, beta 1 |
| chr6_-_5261141 | 19.35 |
ENST00000330636.4
ENST00000500576.2 |
LYRM4
|
LYR motif containing 4 |
| chr2_-_27545921 | 18.98 |
ENST00000402310.1
ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr1_-_86174065 | 18.38 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr6_-_5260963 | 17.71 |
ENST00000464010.1
ENST00000468929.1 ENST00000480566.1 |
LYRM4
|
LYR motif containing 4 |
| chr10_-_97416400 | 17.16 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr8_-_99129338 | 16.72 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chrX_-_16887963 | 16.55 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr5_+_36152091 | 16.47 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr4_-_100009856 | 16.18 |
ENST00000296412.8
|
ADH5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr8_-_48872686 | 15.37 |
ENST00000314191.2
ENST00000338368.3 |
PRKDC
|
protein kinase, DNA-activated, catalytic polypeptide |
| chr22_-_43411106 | 14.25 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr19_-_45953983 | 14.22 |
ENST00000592083.1
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr17_+_36908984 | 14.15 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chrX_-_47518498 | 14.14 |
ENST00000335890.2
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr5_+_36152163 | 13.84 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr21_-_27107344 | 13.44 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr12_-_58165870 | 13.34 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr21_-_27107881 | 13.17 |
ENST00000400090.3
ENST00000400087.3 ENST00000400093.3 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chrX_-_47518527 | 13.14 |
ENST00000333119.3
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr5_-_71616043 | 13.11 |
ENST00000508863.2
ENST00000522095.1 ENST00000513900.1 ENST00000515404.1 ENST00000457646.4 ENST00000261413.5 |
MRPS27
|
mitochondrial ribosomal protein S27 |
| chr1_-_200589859 | 12.41 |
ENST00000367350.4
|
KIF14
|
kinesin family member 14 |
| chr19_-_5719860 | 12.31 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr21_-_27107283 | 12.26 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr22_-_30987849 | 12.17 |
ENST00000402284.3
ENST00000354694.7 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr2_+_207630081 | 11.89 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr2_-_136743169 | 11.68 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr2_+_201676908 | 11.58 |
ENST00000409226.1
ENST00000452790.2 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr17_-_53046058 | 11.37 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr11_-_65626753 | 11.15 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr11_+_6624970 | 10.88 |
ENST00000420936.2
ENST00000528995.1 |
ILK
|
integrin-linked kinase |
| chr12_-_122710556 | 10.84 |
ENST00000446652.1
ENST00000541273.1 ENST00000353548.6 ENST00000267169.6 ENST00000464942.2 |
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr11_+_6624955 | 10.75 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr22_-_30987837 | 10.26 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr11_+_6625046 | 10.21 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chr3_-_98312548 | 9.80 |
ENST00000264193.2
|
CPOX
|
coproporphyrinogen oxidase |
| chr2_+_74154032 | 9.77 |
ENST00000356837.6
|
DGUOK
|
deoxyguanosine kinase |
| chr2_+_74153953 | 9.71 |
ENST00000264093.4
ENST00000348222.1 |
DGUOK
|
deoxyguanosine kinase |
| chr11_-_61560053 | 9.64 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr8_-_99129384 | 9.01 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr9_+_706842 | 8.40 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr14_+_77924373 | 8.39 |
ENST00000216479.3
ENST00000535854.2 ENST00000555517.1 |
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr3_+_133293278 | 8.19 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr20_+_23331373 | 8.13 |
ENST00000254998.2
|
NXT1
|
NTF2-like export factor 1 |
| chr19_-_55652290 | 8.06 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr2_-_112642267 | 7.24 |
ENST00000341068.3
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr2_+_190649107 | 6.80 |
ENST00000441310.2
ENST00000409985.1 ENST00000446877.1 ENST00000418224.3 ENST00000409823.3 ENST00000374826.4 ENST00000424766.1 ENST00000447232.2 ENST00000432292.3 |
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr8_+_109455845 | 6.58 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr13_-_38172863 | 5.87 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr10_+_101491968 | 4.93 |
ENST00000370476.5
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr1_+_118472343 | 4.72 |
ENST00000369441.3
ENST00000349139.5 |
WDR3
|
WD repeat domain 3 |
| chr4_-_57301748 | 4.63 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr12_-_25102252 | 4.63 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr17_-_74733404 | 4.61 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr5_+_89770696 | 4.55 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr6_+_44215603 | 4.53 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr5_+_89770664 | 4.32 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr1_+_231376941 | 3.66 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr12_-_6677422 | 3.61 |
ENST00000382421.3
ENST00000545200.1 ENST00000399466.2 ENST00000536124.1 ENST00000540228.1 ENST00000542867.1 ENST00000545492.1 ENST00000322166.5 ENST00000545915.1 |
NOP2
|
NOP2 nucleolar protein |
| chr16_-_56701933 | 3.59 |
ENST00000568675.1
ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G
|
metallothionein 1G |
| chr17_+_72199721 | 3.47 |
ENST00000439590.2
ENST00000311111.6 ENST00000584577.1 ENST00000534490.1 ENST00000528433.2 ENST00000533498.1 |
RPL38
|
ribosomal protein L38 |
| chr15_+_58724184 | 3.41 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr22_-_50964558 | 3.35 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr16_+_29911864 | 3.22 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr7_+_141463897 | 2.98 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr21_-_30391636 | 2.89 |
ENST00000493196.1
|
RWDD2B
|
RWD domain containing 2B |
| chr8_-_11710979 | 2.85 |
ENST00000415599.2
|
CTSB
|
cathepsin B |
| chr1_+_207070775 | 2.81 |
ENST00000391929.3
ENST00000294984.2 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chr6_-_109804412 | 2.73 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chrX_+_129040094 | 2.68 |
ENST00000425117.2
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr10_-_35379524 | 2.66 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chrX_+_129040122 | 2.63 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr10_-_35379241 | 2.60 |
ENST00000374748.1
ENST00000374749.3 |
CUL2
|
cullin 2 |
| chr10_-_105677427 | 2.55 |
ENST00000369764.1
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr17_-_13505219 | 2.16 |
ENST00000284110.1
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr12_-_25101920 | 2.08 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr6_+_5261225 | 1.99 |
ENST00000324331.6
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr17_+_48796905 | 1.64 |
ENST00000505658.1
ENST00000393227.2 ENST00000240304.1 ENST00000311571.3 ENST00000505619.1 ENST00000544170.1 ENST00000510984.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr15_-_74988281 | 1.44 |
ENST00000566828.1
ENST00000563009.1 ENST00000568176.1 ENST00000566243.1 ENST00000566219.1 ENST00000426797.3 ENST00000566119.1 ENST00000315127.4 |
EDC3
|
enhancer of mRNA decapping 3 |
| chr4_+_47487285 | 1.27 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr1_+_44445549 | 0.90 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr8_+_32405728 | 0.82 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr5_-_114880533 | 0.80 |
ENST00000274457.3
|
FEM1C
|
fem-1 homolog c (C. elegans) |
| chr1_-_113498943 | 0.57 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr19_+_41497178 | 0.25 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr7_-_151574191 | 0.19 |
ENST00000287878.4
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE40
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.1 | 90.6 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 17.8 | 53.3 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 14.7 | 58.9 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 8.1 | 24.3 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 5.9 | 59.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 5.8 | 29.0 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 5.4 | 16.2 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 4.9 | 19.6 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 4.9 | 19.5 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 4.1 | 12.3 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 3.3 | 16.6 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 3.1 | 18.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 2.9 | 17.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 2.8 | 11.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 2.8 | 8.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 2.5 | 12.4 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 2.4 | 11.9 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 2.4 | 14.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 2.3 | 6.8 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 2.2 | 22.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 2.2 | 10.8 | GO:0071964 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 2.0 | 22.0 | GO:0006983 | ER overload response(GO:0006983) |
| 2.0 | 5.9 | GO:1990523 | bone regeneration(GO:1990523) |
| 1.9 | 15.4 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) ectopic germ cell programmed cell death(GO:0035234) |
| 1.4 | 14.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 1.3 | 31.8 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 1.3 | 96.7 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 1.2 | 3.7 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 1.2 | 38.9 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.2 | 19.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 1.2 | 39.5 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 1.1 | 4.5 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 1.1 | 6.7 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 1.1 | 27.3 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 1.0 | 9.8 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.9 | 8.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.9 | 66.3 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.7 | 10.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.5 | 2.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.5 | 46.5 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.4 | 6.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 3.6 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.4 | 4.6 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.3 | 8.3 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.3 | 9.8 | GO:0044764 | viral process(GO:0016032) multi-organism cellular process(GO:0044764) |
| 0.3 | 7.9 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.2 | 3.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 2.2 | GO:0006023 | aminoglycan metabolic process(GO:0006022) aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) glycosaminoglycan metabolic process(GO:0030203) |
| 0.2 | 13.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.2 | 9.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.2 | 1.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 21.4 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.1 | 0.6 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 2.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 2.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 3.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 5.3 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 2.8 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 8.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 8.1 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.3 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.9 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 1.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.6 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 4.9 | 39.5 | GO:0005683 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 3.7 | 22.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 2.7 | 13.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 2.6 | 15.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 2.4 | 59.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 2.3 | 18.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 2.2 | 24.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 2.2 | 38.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 2.1 | 83.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 2.0 | 14.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 2.0 | 87.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 1.1 | 4.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 1.1 | 5.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.9 | 4.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.9 | 4.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.9 | 14.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.9 | 6.8 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.8 | 9.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.8 | 10.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.7 | 6.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.6 | 31.8 | GO:0043034 | costamere(GO:0043034) |
| 0.6 | 16.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.6 | 46.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.6 | 15.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.5 | 29.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.4 | 24.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 2.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.4 | 12.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.4 | 8.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.3 | 8.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 6.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 10.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 7.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 20.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 14.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 50.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 65.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 3.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 3.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 8.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 8.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 4.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 3.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 90.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 8.1 | 24.3 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 5.1 | 15.4 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 4.3 | 17.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 4.1 | 12.3 | GO:0070364 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 3.9 | 39.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 3.2 | 19.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 2.4 | 54.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 2.2 | 32.6 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 1.6 | 16.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 1.5 | 4.5 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 1.2 | 3.6 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 1.1 | 6.7 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.1 | 13.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 1.1 | 58.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 1.0 | 6.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 1.0 | 53.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.8 | 9.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.6 | 27.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.6 | 14.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.6 | 8.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.5 | 2.0 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.5 | 14.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 12.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 11.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 37.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.4 | 87.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.4 | 2.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 3.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) O-acetyltransferase activity(GO:0016413) |
| 0.3 | 16.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.3 | 3.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.3 | 3.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.3 | 8.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 0.9 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 4.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.2 | 17.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 8.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 8.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.2 | 31.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 3.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 34.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 3.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 10.8 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.1 | 6.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 4.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 4.6 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 2.9 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 4.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 7.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 102.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.1 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 5.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 17.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.9 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 2.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 19.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.8 | 25.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.6 | 29.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.6 | 65.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.6 | 19.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.5 | 15.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.5 | 37.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 25.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 16.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 5.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 10.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 4.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 5.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 59.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 2.5 | 39.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 2.4 | 58.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 1.7 | 38.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 1.1 | 31.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.8 | 19.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.8 | 47.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.6 | 17.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.5 | 15.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.5 | 14.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.5 | 15.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.5 | 50.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.4 | 9.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 15.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 6.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 6.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 5.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 3.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 19.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 8.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 4.6 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 3.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |