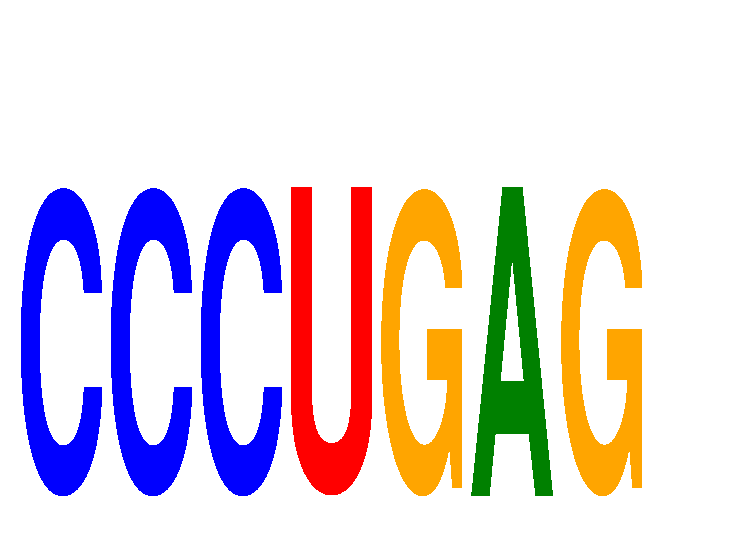Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for CCCUGAG
Z-value: 1.86
miRNA associated with seed CCCUGAG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-125a-5p
|
MIMAT0000443 |
|
hsa-miR-125b-5p
|
MIMAT0000423 |
|
hsa-miR-4319
|
MIMAT0016870 |
Activity profile of CCCUGAG motif
Sorted Z-values of CCCUGAG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_64708615 | 37.28 |
ENST00000338957.4
ENST00000423889.3 |
ZC3H12B
|
zinc finger CCCH-type containing 12B |
| chr15_+_31619013 | 30.24 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr22_-_39548627 | 23.17 |
ENST00000216133.5
|
CBX7
|
chromobox homolog 7 |
| chr15_+_74833518 | 21.55 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr9_-_116102530 | 21.04 |
ENST00000374195.3
ENST00000341761.4 |
WDR31
|
WD repeat domain 31 |
| chrX_-_103401649 | 20.71 |
ENST00000357421.4
|
SLC25A53
|
solute carrier family 25, member 53 |
| chr4_-_2264015 | 18.86 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr2_-_206950781 | 18.74 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr17_-_61777459 | 18.59 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr19_+_7968728 | 17.92 |
ENST00000397981.3
ENST00000545011.1 ENST00000397983.3 ENST00000397979.3 |
MAP2K7
|
mitogen-activated protein kinase kinase 7 |
| chr3_-_57678772 | 17.40 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr3_+_15468862 | 17.28 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chr16_+_1662326 | 16.64 |
ENST00000397412.3
|
CRAMP1L
|
Crm, cramped-like (Drosophila) |
| chr19_+_18794470 | 15.32 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr19_-_18632861 | 15.15 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr16_-_28936493 | 14.98 |
ENST00000544477.1
ENST00000357573.6 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr22_+_41777927 | 14.55 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr7_-_139876812 | 14.46 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr17_-_76713100 | 14.14 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr2_+_61293021 | 13.57 |
ENST00000402291.1
|
KIAA1841
|
KIAA1841 |
| chr2_-_201936302 | 13.51 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr9_-_123476719 | 13.32 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr10_+_120967072 | 13.32 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr6_+_106546808 | 13.22 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr16_-_2264779 | 12.87 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr22_-_44258360 | 12.81 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr1_+_203274639 | 12.71 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr11_-_62494821 | 12.62 |
ENST00000301785.5
|
HNRNPUL2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr4_+_7045042 | 12.57 |
ENST00000310074.7
ENST00000512388.1 |
TADA2B
|
transcriptional adaptor 2B |
| chr1_+_12227035 | 12.55 |
ENST00000376259.3
ENST00000536782.1 |
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B |
| chr3_+_47021168 | 12.54 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chrX_+_110339439 | 12.40 |
ENST00000372010.1
ENST00000519681.1 ENST00000372007.5 |
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr5_-_159739532 | 12.36 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr3_+_183353356 | 12.34 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr11_-_117103208 | 12.23 |
ENST00000320934.3
ENST00000530269.1 |
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr11_-_22851367 | 12.18 |
ENST00000354193.4
|
SVIP
|
small VCP/p97-interacting protein |
| chr1_-_207224307 | 12.04 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr3_+_186648274 | 12.02 |
ENST00000169298.3
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr16_+_4897632 | 11.90 |
ENST00000262376.6
|
UBN1
|
ubinuclein 1 |
| chr6_+_138188551 | 11.43 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr12_+_11802753 | 11.34 |
ENST00000396373.4
|
ETV6
|
ets variant 6 |
| chr1_+_78245303 | 11.26 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr9_-_3525968 | 11.19 |
ENST00000382004.3
ENST00000302303.1 ENST00000449190.1 |
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr9_-_127533519 | 11.16 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr6_+_391739 | 10.92 |
ENST00000380956.4
|
IRF4
|
interferon regulatory factor 4 |
| chr15_-_72410109 | 10.87 |
ENST00000564571.1
|
MYO9A
|
myosin IXA |
| chr1_-_212208842 | 10.80 |
ENST00000366992.3
ENST00000366993.3 ENST00000440600.2 ENST00000366994.3 |
INTS7
|
integrator complex subunit 7 |
| chr5_-_175964366 | 10.67 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr3_+_50712672 | 10.53 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr7_+_49813255 | 10.32 |
ENST00000340652.4
|
VWC2
|
von Willebrand factor C domain containing 2 |
| chr17_-_36956155 | 10.11 |
ENST00000269554.3
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr6_+_138483058 | 10.05 |
ENST00000251691.4
|
KIAA1244
|
KIAA1244 |
| chr17_+_42836329 | 9.98 |
ENST00000200557.6
|
ADAM11
|
ADAM metallopeptidase domain 11 |
| chr15_-_90358048 | 9.96 |
ENST00000300060.6
ENST00000560137.1 |
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr15_+_81489213 | 9.89 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr10_-_15762124 | 9.88 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr3_+_49591881 | 9.75 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr15_-_61521495 | 9.72 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chrX_+_153639856 | 9.57 |
ENST00000426834.1
ENST00000369790.4 ENST00000454722.1 ENST00000350743.4 ENST00000299328.5 ENST00000351413.4 |
TAZ
|
tafazzin |
| chr16_+_27561449 | 9.41 |
ENST00000261588.4
|
KIAA0556
|
KIAA0556 |
| chr16_-_18937726 | 9.40 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr4_-_25032501 | 9.34 |
ENST00000382114.4
|
LGI2
|
leucine-rich repeat LGI family, member 2 |
| chr7_-_143059845 | 9.29 |
ENST00000443739.2
|
FAM131B
|
family with sequence similarity 131, member B |
| chr14_+_69865401 | 9.22 |
ENST00000556605.1
ENST00000336643.5 ENST00000031146.4 |
SLC39A9
|
solute carrier family 39, member 9 |
| chr2_+_148602058 | 9.20 |
ENST00000241416.7
ENST00000535787.1 ENST00000404590.1 |
ACVR2A
|
activin A receptor, type IIA |
| chr16_-_71610985 | 9.14 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr3_-_13461807 | 9.01 |
ENST00000254508.5
|
NUP210
|
nucleoporin 210kDa |
| chr19_+_40697514 | 8.54 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr2_-_38604398 | 8.48 |
ENST00000443098.1
ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2
|
atlastin GTPase 2 |
| chr6_-_24911195 | 8.38 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr11_-_46940074 | 8.35 |
ENST00000378623.1
ENST00000534404.1 |
LRP4
|
low density lipoprotein receptor-related protein 4 |
| chr12_-_120806960 | 8.33 |
ENST00000257552.2
|
MSI1
|
musashi RNA-binding protein 1 |
| chr5_+_56111361 | 8.30 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr3_+_141205852 | 8.14 |
ENST00000286364.3
ENST00000452898.1 |
RASA2
|
RAS p21 protein activator 2 |
| chr11_+_33278811 | 8.09 |
ENST00000303296.4
ENST00000379016.3 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr10_+_101088836 | 8.09 |
ENST00000356713.4
|
CNNM1
|
cyclin M1 |
| chr10_+_104678032 | 8.05 |
ENST00000369878.4
ENST00000369875.3 |
CNNM2
|
cyclin M2 |
| chr19_+_19322758 | 8.02 |
ENST00000252575.6
|
NCAN
|
neurocan |
| chr3_+_37493610 | 8.02 |
ENST00000264741.5
|
ITGA9
|
integrin, alpha 9 |
| chr14_+_74111578 | 8.00 |
ENST00000554113.1
ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1
|
dynein, axonemal, light chain 1 |
| chr11_-_118047376 | 7.91 |
ENST00000278947.5
|
SCN2B
|
sodium channel, voltage-gated, type II, beta subunit |
| chr1_+_32573636 | 7.85 |
ENST00000373625.3
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr12_+_56401268 | 7.85 |
ENST00000262032.5
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr19_+_45596218 | 7.83 |
ENST00000421905.1
ENST00000221462.4 |
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr5_+_112043186 | 7.79 |
ENST00000509732.1
ENST00000457016.1 ENST00000507379.1 |
APC
|
adenomatous polyposis coli |
| chr20_-_4982132 | 7.78 |
ENST00000338244.1
ENST00000424750.2 |
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr1_-_109940550 | 7.70 |
ENST00000256637.6
|
SORT1
|
sortilin 1 |
| chr11_-_119252359 | 7.60 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr12_-_42538657 | 7.58 |
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr10_+_82168240 | 7.53 |
ENST00000372187.5
ENST00000372185.1 |
FAM213A
|
family with sequence similarity 213, member A |
| chr18_+_32558208 | 7.34 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_-_21671968 | 7.33 |
ENST00000415912.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr20_+_55966444 | 7.31 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr14_-_100070363 | 7.30 |
ENST00000380243.4
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr2_-_103353277 | 7.25 |
ENST00000258436.5
|
MFSD9
|
major facilitator superfamily domain containing 9 |
| chr1_-_36022979 | 7.20 |
ENST00000469892.1
ENST00000325722.3 |
KIAA0319L
|
KIAA0319-like |
| chr19_-_4066890 | 7.12 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr1_-_211307315 | 7.11 |
ENST00000271751.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr7_+_142985308 | 7.08 |
ENST00000310447.5
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr2_-_9143786 | 7.07 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr12_-_56727487 | 7.02 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr19_-_47975417 | 6.99 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr1_-_179198702 | 6.94 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr9_-_130829588 | 6.93 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr12_+_13197218 | 6.74 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr3_-_133614597 | 6.66 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr9_+_35538616 | 6.65 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr16_+_68298405 | 6.64 |
ENST00000219343.6
ENST00000566834.1 ENST00000566454.1 |
SLC7A6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr11_+_94277017 | 6.63 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr8_-_67579418 | 6.61 |
ENST00000310421.4
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr12_+_5019061 | 6.56 |
ENST00000382545.3
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) |
| chr16_-_75498553 | 6.43 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr2_-_213403565 | 6.38 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr15_+_68871308 | 6.38 |
ENST00000261861.5
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr13_-_73356009 | 6.34 |
ENST00000377780.4
ENST00000377767.4 |
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr15_-_73661605 | 6.31 |
ENST00000261917.3
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr17_-_48207157 | 6.22 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr3_-_53381539 | 6.11 |
ENST00000606822.1
ENST00000294241.6 ENST00000607628.1 |
DCP1A
|
decapping mRNA 1A |
| chr14_+_56585048 | 6.08 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr3_+_58223228 | 6.05 |
ENST00000478253.1
ENST00000295962.4 |
ABHD6
|
abhydrolase domain containing 6 |
| chr1_-_23857698 | 6.02 |
ENST00000361729.2
|
E2F2
|
E2F transcription factor 2 |
| chr17_-_66287257 | 5.96 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr10_-_50747064 | 5.95 |
ENST00000355832.5
ENST00000603152.1 ENST00000447839.2 |
ERCC6
PGBD3
ERCC6-PGBD3
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 piggyBac transposable element derived 3 ERCC6-PGBD3 readthrough |
| chr10_+_102295616 | 5.94 |
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr15_-_79383102 | 5.93 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr19_-_49944806 | 5.87 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr7_-_98741642 | 5.87 |
ENST00000361368.2
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr10_+_73975742 | 5.79 |
ENST00000299381.4
|
ANAPC16
|
anaphase promoting complex subunit 16 |
| chr6_-_100912785 | 5.79 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr1_+_205197304 | 5.78 |
ENST00000358024.3
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chrX_+_24483338 | 5.76 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr3_+_5229356 | 5.75 |
ENST00000256497.4
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr22_+_40390930 | 5.74 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr5_+_96271141 | 5.74 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chr17_+_41177220 | 5.59 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr22_-_50699701 | 5.58 |
ENST00000395780.1
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr19_-_19754404 | 5.57 |
ENST00000587205.1
ENST00000445806.2 ENST00000203556.4 |
GMIP
|
GEM interacting protein |
| chr13_-_29292956 | 5.54 |
ENST00000266943.6
|
SLC46A3
|
solute carrier family 46, member 3 |
| chr19_+_4969116 | 5.48 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr2_-_50574856 | 5.47 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr16_+_8768422 | 5.40 |
ENST00000268251.8
ENST00000564714.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr17_-_27224621 | 5.40 |
ENST00000394906.2
ENST00000585169.1 ENST00000394908.4 |
FLOT2
|
flotillin 2 |
| chr8_-_63998590 | 5.23 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr1_+_26856236 | 5.23 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr9_-_95432536 | 5.17 |
ENST00000287996.3
|
IPPK
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr9_-_35650900 | 5.13 |
ENST00000259608.3
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr20_+_3827459 | 5.12 |
ENST00000416600.2
ENST00000428216.2 |
MAVS
|
mitochondrial antiviral signaling protein |
| chr17_-_42908155 | 5.12 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr11_-_128392085 | 5.11 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr10_+_99258625 | 5.08 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr18_+_55102917 | 4.99 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr8_+_22857048 | 4.99 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr17_+_61699766 | 4.93 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr17_-_8093471 | 4.92 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr10_-_94003003 | 4.87 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr9_-_19786926 | 4.80 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr12_-_69326940 | 4.60 |
ENST00000549781.1
ENST00000548262.1 ENST00000551568.1 ENST00000548954.1 |
CPM
|
carboxypeptidase M |
| chr6_+_35227449 | 4.59 |
ENST00000373953.3
ENST00000440666.2 ENST00000339411.5 |
ZNF76
|
zinc finger protein 76 |
| chr11_+_71640071 | 4.58 |
ENST00000533380.1
ENST00000393713.3 ENST00000545854.1 |
RNF121
|
ring finger protein 121 |
| chr12_-_51717922 | 4.56 |
ENST00000452142.2
|
BIN2
|
bridging integrator 2 |
| chr3_-_183735731 | 4.56 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr8_-_145550571 | 4.54 |
ENST00000332324.4
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr11_+_58939965 | 4.54 |
ENST00000227451.3
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr12_+_53774423 | 4.48 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr19_-_11450249 | 4.47 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr2_+_18059906 | 4.46 |
ENST00000304101.4
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr5_-_141257954 | 4.28 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr14_-_92572894 | 4.26 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr1_-_150552006 | 4.22 |
ENST00000307940.3
ENST00000369026.2 |
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related) |
| chr1_-_157108130 | 4.14 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chr1_-_200379180 | 4.11 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr5_-_100238956 | 4.08 |
ENST00000231461.5
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr17_-_5372271 | 4.06 |
ENST00000225296.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr10_+_14920843 | 4.06 |
ENST00000433779.1
ENST00000378325.3 ENST00000354919.6 ENST00000313519.5 ENST00000420416.1 |
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila) |
| chr1_-_23495340 | 4.06 |
ENST00000418342.1
|
LUZP1
|
leucine zipper protein 1 |
| chr5_-_37839782 | 4.06 |
ENST00000326524.2
ENST00000515058.1 |
GDNF
|
glial cell derived neurotrophic factor |
| chr4_+_37455536 | 4.02 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr20_+_5107420 | 3.98 |
ENST00000460006.1
|
CDS2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr3_-_170626418 | 3.94 |
ENST00000474096.1
ENST00000295822.2 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr20_-_48099182 | 3.94 |
ENST00000371741.4
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr4_+_72204755 | 3.93 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr14_-_82000140 | 3.92 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr12_+_69633317 | 3.91 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr22_+_21771656 | 3.88 |
ENST00000407464.2
|
HIC2
|
hypermethylated in cancer 2 |
| chr1_+_151584544 | 3.87 |
ENST00000458013.2
ENST00000368843.3 |
SNX27
|
sorting nexin family member 27 |
| chr9_-_100935043 | 3.87 |
ENST00000343933.5
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr15_+_85923856 | 3.84 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr6_-_90062543 | 3.83 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr17_-_40333099 | 3.81 |
ENST00000607371.1
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr3_+_38495333 | 3.79 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr8_-_8751068 | 3.77 |
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr15_-_93199069 | 3.74 |
ENST00000327355.5
|
FAM174B
|
family with sequence similarity 174, member B |
| chr4_+_38665810 | 3.72 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr18_-_60987220 | 3.67 |
ENST00000398117.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr9_+_79074068 | 3.63 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr10_+_82213904 | 3.61 |
ENST00000429989.3
|
TSPAN14
|
tetraspanin 14 |
| chr16_-_23521710 | 3.58 |
ENST00000562117.1
ENST00000567468.1 ENST00000562944.1 ENST00000309859.4 |
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr3_+_179065474 | 3.56 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr1_+_178062855 | 3.54 |
ENST00000448150.3
|
RASAL2
|
RAS protein activator like 2 |
| chr2_+_177053307 | 3.52 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CCCUGAG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 4.1 | 12.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 4.0 | 12.0 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 3.9 | 19.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 3.8 | 15.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 3.7 | 11.2 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 3.6 | 10.9 | GO:0045082 | interleukin-4 biosynthetic process(GO:0042097) positive regulation of interleukin-10 biosynthetic process(GO:0045082) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 3.2 | 12.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 3.2 | 6.4 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 3.0 | 30.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 2.8 | 8.3 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 2.6 | 13.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 2.4 | 9.6 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 2.2 | 17.9 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 2.1 | 6.3 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 2.0 | 5.9 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 2.0 | 3.9 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 2.0 | 5.9 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 2.0 | 5.9 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 1.9 | 13.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.9 | 11.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.8 | 7.3 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.7 | 12.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.6 | 4.9 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 1.6 | 17.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 1.6 | 10.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.5 | 9.2 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 1.5 | 6.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.4 | 5.4 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 1.3 | 4.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 1.3 | 7.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.3 | 5.2 | GO:0090210 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 1.3 | 9.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.3 | 3.8 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.2 | 6.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.1 | 5.7 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.1 | 4.5 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 1.1 | 7.8 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 1.1 | 2.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 1.1 | 4.3 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 1.1 | 6.3 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 1.0 | 1.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 1.0 | 7.3 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 1.0 | 3.1 | GO:0001743 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 1.0 | 5.1 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) regulation of peroxisome organization(GO:1900063) |
| 1.0 | 5.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 1.0 | 4.1 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 1.0 | 7.1 | GO:0001554 | luteolysis(GO:0001554) |
| 1.0 | 3.0 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 1.0 | 3.0 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 1.0 | 10.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 1.0 | 3.8 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.9 | 6.6 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.9 | 6.5 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.9 | 12.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.9 | 3.7 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.9 | 12.7 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.9 | 5.4 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.9 | 4.5 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.9 | 5.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.9 | 6.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.8 | 10.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.8 | 5.8 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.8 | 3.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.8 | 7.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.8 | 3.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.7 | 9.7 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.7 | 4.5 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.7 | 3.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.7 | 2.2 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.7 | 14.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.7 | 2.1 | GO:1905075 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.7 | 4.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.7 | 5.5 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.7 | 5.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.7 | 8.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.7 | 6.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.7 | 2.0 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.7 | 3.3 | GO:2000035 | positive regulation of somatic stem cell population maintenance(GO:1904674) regulation of stem cell division(GO:2000035) |
| 0.7 | 3.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.6 | 5.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.6 | 7.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.6 | 1.9 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.6 | 0.6 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.6 | 4.8 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.6 | 8.1 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.6 | 5.7 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.6 | 4.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.6 | 1.7 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.6 | 3.9 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.5 | 2.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.5 | 1.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.5 | 1.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.5 | 12.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.5 | 3.6 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.5 | 12.4 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.5 | 4.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.5 | 3.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.5 | 3.9 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 | 2.4 | GO:0090625 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.5 | 1.4 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.5 | 7.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.5 | 9.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.5 | 2.4 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.5 | 1.4 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.5 | 6.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.4 | 3.0 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.4 | 5.0 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.4 | 14.7 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.4 | 8.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.4 | 12.8 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.4 | 1.2 | GO:0046016 | carbon catabolite activation of transcription(GO:0045991) positive regulation of transcription by glucose(GO:0046016) |
| 0.4 | 4.1 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.4 | 5.6 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.4 | 9.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.4 | 2.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.3 | 8.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.3 | 12.6 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.3 | 9.0 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.3 | 12.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.3 | 4.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.3 | 2.7 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.3 | 5.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.3 | 1.6 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.3 | 5.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 8.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 | 1.3 | GO:0051944 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.3 | 1.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 3.8 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.3 | 0.9 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.3 | 10.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.3 | 4.9 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.3 | 4.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.3 | 7.0 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.3 | 8.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.3 | 0.5 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 | 11.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.2 | 3.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.2 | 3.0 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.2 | 6.1 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.2 | 0.9 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.7 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) dense core granule biogenesis(GO:0061110) negative regulation of cortisol biosynthetic process(GO:2000065) regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 3.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 3.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 10.8 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 0.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 1.8 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.2 | 3.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.2 | 1.4 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.2 | 0.8 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.2 | 11.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.2 | 1.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 3.4 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.2 | 4.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 4.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.2 | 5.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.9 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 3.0 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 | 4.6 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.2 | 9.0 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.2 | 5.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.2 | 6.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 4.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 6.6 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.2 | 5.8 | GO:1904667 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition(GO:0051437) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 2.3 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.2 | 12.8 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.2 | 6.9 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.2 | 9.8 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.2 | 2.4 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 7.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.7 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 2.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 5.7 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.1 | 2.3 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 1.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 2.9 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.1 | 2.9 | GO:0009584 | detection of visible light(GO:0009584) |
| 0.1 | 4.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 6.8 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.1 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 7.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 7.0 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 2.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.4 | GO:0048014 | Tie signaling pathway(GO:0048014) negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 3.1 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 2.8 | GO:0003338 | metanephros morphogenesis(GO:0003338) |
| 0.1 | 1.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 2.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.6 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 1.4 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.1 | 1.6 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.1 | 1.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 3.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 3.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.8 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 1.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 17.6 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.1 | 3.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.9 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 16.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 4.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 6.8 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 7.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 2.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.7 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.6 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 2.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 2.7 | GO:0016032 | viral process(GO:0016032) multi-organism cellular process(GO:0044764) |
| 0.1 | 6.6 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 4.5 | GO:0032479 | regulation of type I interferon production(GO:0032479) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 3.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 7.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 3.5 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 3.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 6.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 2.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 2.9 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.7 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.4 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 4.5 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 1.6 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 7.0 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.7 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 1.1 | GO:0072164 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.1 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.9 | GO:0070997 | neuron death(GO:0070997) |
| 0.0 | 0.8 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 1.2 | GO:0006310 | DNA recombination(GO:0006310) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 2.3 | 7.0 | GO:0031251 | PAN complex(GO:0031251) |
| 2.3 | 9.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.8 | 7.3 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 1.7 | 23.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 1.6 | 13.2 | GO:0043196 | varicosity(GO:0043196) |
| 1.5 | 16.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 1.4 | 5.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 1.2 | 10.8 | GO:0032039 | integrator complex(GO:0032039) |
| 1.1 | 5.7 | GO:0031905 | early endosome lumen(GO:0031905) |
| 1.0 | 8.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.9 | 10.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.8 | 3.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.8 | 12.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.8 | 10.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.7 | 7.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.7 | 5.9 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.7 | 10.9 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.7 | 38.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.6 | 7.8 | GO:1990909 | catenin complex(GO:0016342) Wnt signalosome(GO:1990909) |
| 0.6 | 11.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.6 | 6.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.6 | 2.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.6 | 3.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.5 | 1.6 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.5 | 3.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.5 | 3.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 2.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 6.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.5 | 7.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 1.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.4 | 11.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 15.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.4 | 3.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.4 | 2.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.3 | 3.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 12.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.3 | 4.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 5.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 1.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 7.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 23.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 2.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 28.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 4.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 34.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 27.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 4.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 9.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 2.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 2.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.2 | 5.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 12.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 2.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 0.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 4.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 12.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 2.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 6.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.1 | 2.7 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 9.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 7.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 16.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 13.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 10.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 3.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 19.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 5.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 1.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 30.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 16.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 2.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 5.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 11.8 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 2.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 25.2 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 5.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 6.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.6 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 2.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 12.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 5.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 9.9 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 4.3 | 12.9 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 4.0 | 12.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 4.0 | 12.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 3.4 | 10.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 3.0 | 9.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 2.6 | 7.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 2.3 | 11.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 2.0 | 15.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.9 | 13.0 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 1.8 | 10.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.8 | 7.1 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 1.6 | 7.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.6 | 6.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.6 | 6.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.5 | 7.7 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 1.5 | 7.6 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 1.5 | 4.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.5 | 5.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.4 | 5.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.4 | 9.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.4 | 5.4 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 1.3 | 4.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 1.3 | 5.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 1.2 | 3.6 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 1.1 | 30.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 1.0 | 5.1 | GO:0086077 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 1.0 | 15.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 1.0 | 4.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 1.0 | 4.8 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.8 | 5.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.8 | 6.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.8 | 18.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.8 | 3.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.8 | 2.3 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.8 | 3.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.7 | 2.2 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.7 | 7.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.7 | 7.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.7 | 5.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.7 | 2.1 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.7 | 15.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.7 | 3.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.6 | 1.8 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.6 | 4.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.6 | 8.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.6 | 5.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.6 | 12.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.5 | 14.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.5 | 2.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.5 | 6.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.5 | 1.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.5 | 5.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.5 | 1.9 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.5 | 6.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.5 | 4.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.4 | 9.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 21.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.4 | 13.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 11.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 6.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.4 | 12.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 8.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 7.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.4 | 7.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 3.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.4 | 4.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.4 | 6.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 5.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 4.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.4 | 13.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.4 | 2.1 | GO:0005114 | transforming growth factor beta receptor activity, type I(GO:0005025) type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.4 | 18.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 5.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 1.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 6.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.3 | 3.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 6.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.3 | 4.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 0.9 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 4.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 4.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.3 | 12.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.3 | 7.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.3 | 8.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 2.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 1.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 5.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.3 | 17.2 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.3 | 12.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 1.7 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 3.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 15.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 3.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 7.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 3.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 10.9 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 1.8 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.2 | 2.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 17.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 3.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 2.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 2.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 4.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 25.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 7.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 2.8 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.2 | 0.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 7.3 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 6.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 5.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 9.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 4.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 18.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 2.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 1.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 8.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 36.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 6.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 12.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 6.6 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 3.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 5.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 6.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 5.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 0.9 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.9 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.7 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 2.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 28.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 8.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 3.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 2.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 2.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 5.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 3.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 12.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 13.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.2 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 3.2 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 8.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 10.9 | GO:0022857 | transmembrane transporter activity(GO:0022857) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 56.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.4 | 14.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 5.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 8.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 14.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 15.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 12.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 8.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 3.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.3 | 10.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.3 | 6.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 7.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 2.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 9.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 2.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 7.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 14.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 4.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 10.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 6.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 3.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 8.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 14.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 5.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 6.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 14.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 10.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 3.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 37.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 6.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 6.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 4.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 15.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 19.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.9 | 6.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.8 | 23.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.7 | 6.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.6 | 12.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.6 | 6.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.6 | 5.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 25.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.5 | 5.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.5 | 18.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.5 | 12.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 2.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 9.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.3 | 9.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 8.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.3 | 15.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.3 | 5.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 7.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 2.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 6.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 8.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 12.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.3 | 5.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 5.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.3 | 10.8 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.3 | 6.0 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.3 | 5.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 4.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 7.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 5.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.2 | 5.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 17.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 18.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.2 | 12.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.2 | 6.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 5.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 3.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 1.4 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.2 | 6.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 5.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 4.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 11.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 4.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 9.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 6.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 2.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 5.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 3.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 4.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 7.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 5.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 14.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 12.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 6.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 4.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |