Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
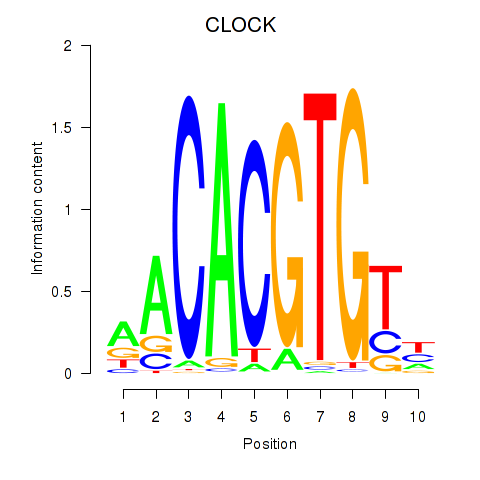
Results for CLOCK
Z-value: 1.70
Transcription factors associated with CLOCK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CLOCK
|
ENSG00000134852.10 | clock circadian regulator |
Activity profile of CLOCK motif
Sorted Z-values of CLOCK motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_64014379 | 41.56 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_-_26233423 | 37.74 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr5_+_138609782 | 35.70 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr17_-_47492164 | 30.72 |
ENST00000512041.2
ENST00000446735.1 ENST00000504124.1 |
PHB
|
prohibitin |
| chr2_+_242289502 | 29.31 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr2_+_198365122 | 28.86 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr12_+_57624085 | 26.81 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_-_58146128 | 26.74 |
ENST00000551800.1
ENST00000549606.1 ENST00000257904.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr12_+_64798095 | 26.28 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr2_+_201170770 | 26.26 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr16_-_81129845 | 26.08 |
ENST00000569885.1
ENST00000566566.1 |
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr20_+_44441304 | 25.90 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr15_-_49447771 | 25.73 |
ENST00000558843.1
ENST00000542928.1 ENST00000561248.1 |
COPS2
|
COP9 signalosome subunit 2 |
| chr12_-_58146048 | 25.40 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr20_+_44441215 | 24.92 |
ENST00000356455.4
ENST00000405520.1 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr4_+_57302297 | 24.80 |
ENST00000399688.3
ENST00000512576.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr1_-_26232951 | 24.77 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr2_+_201170596 | 24.20 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chrX_+_23685653 | 24.07 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr11_+_114310102 | 24.01 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr17_-_48474828 | 23.95 |
ENST00000576448.1
ENST00000225972.7 |
LRRC59
|
leucine rich repeat containing 59 |
| chr2_+_201171242 | 23.77 |
ENST00000360760.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr9_+_112542591 | 23.17 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr2_+_216176761 | 23.00 |
ENST00000540518.1
ENST00000435675.1 |
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr11_-_122931881 | 22.88 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr11_+_18416103 | 22.73 |
ENST00000543445.1
ENST00000430553.2 ENST00000396222.2 ENST00000535451.1 |
LDHA
|
lactate dehydrogenase A |
| chr11_+_114310237 | 22.71 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr5_+_138609441 | 22.58 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr2_+_198365095 | 22.51 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr11_+_114310164 | 22.43 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chrX_+_23685563 | 22.37 |
ENST00000379341.4
|
PRDX4
|
peroxiredoxin 4 |
| chr12_+_57624119 | 22.17 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_-_159894319 | 22.04 |
ENST00000320307.4
|
TAGLN2
|
transgelin 2 |
| chr1_-_11120057 | 21.83 |
ENST00000376957.2
|
SRM
|
spermidine synthase |
| chr4_+_57301896 | 21.15 |
ENST00000514888.1
ENST00000264221.2 ENST00000505164.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr12_-_76477707 | 21.10 |
ENST00000551992.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr17_+_49243639 | 21.03 |
ENST00000512737.1
ENST00000503064.1 |
NME1-NME2
|
NME1-NME2 readthrough |
| chr17_+_40985407 | 20.97 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr11_+_18416133 | 20.82 |
ENST00000227157.4
ENST00000478970.2 ENST00000495052.1 |
LDHA
|
lactate dehydrogenase A |
| chr9_+_112542572 | 20.31 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr14_-_58893832 | 20.28 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr17_-_40075219 | 20.17 |
ENST00000537919.1
ENST00000352035.2 ENST00000353196.1 ENST00000393896.2 |
ACLY
|
ATP citrate lyase |
| chr20_+_44441271 | 20.03 |
ENST00000335046.3
ENST00000243893.6 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr2_+_216176540 | 20.02 |
ENST00000236959.9
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr12_-_21810726 | 19.85 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr17_+_49243792 | 19.49 |
ENST00000393183.3
ENST00000393190.1 |
NME1-NME2
|
NME1-NME2 readthrough |
| chr1_+_84944926 | 19.40 |
ENST00000370656.1
ENST00000370654.5 |
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae) |
| chr12_-_21810765 | 19.17 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr14_-_58894223 | 18.91 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr5_+_34656569 | 18.84 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr5_+_135394840 | 18.56 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr3_+_52719936 | 18.33 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr17_-_5342380 | 18.28 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr2_+_201171372 | 18.24 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr11_+_65770227 | 17.97 |
ENST00000527348.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr12_+_57623869 | 17.51 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_-_94285402 | 17.29 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr19_+_50180317 | 17.14 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr2_-_106013400 | 16.77 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr2_-_175113088 | 16.60 |
ENST00000409546.1
ENST00000428402.2 |
OLA1
|
Obg-like ATPase 1 |
| chr17_-_61850894 | 16.51 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr5_+_34656331 | 16.36 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr7_+_56019486 | 16.29 |
ENST00000446692.1
ENST00000285298.4 ENST00000443449.1 |
GBAS
MRPS17
|
glioblastoma amplified sequence mitochondrial ribosomal protein S17 |
| chr12_+_109535373 | 16.27 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chr10_+_60028818 | 16.08 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr2_+_187350883 | 15.86 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr1_+_166808692 | 15.65 |
ENST00000367876.4
|
POGK
|
pogo transposable element with KRAB domain |
| chr1_+_38478378 | 15.48 |
ENST00000373014.4
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr2_-_47143160 | 15.43 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr6_-_30712313 | 15.29 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr2_-_10587897 | 15.27 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr7_-_44163107 | 15.15 |
ENST00000406581.2
ENST00000452185.1 ENST00000436844.1 ENST00000418438.1 |
POLD2
|
polymerase (DNA directed), delta 2, accessory subunit |
| chr1_+_182992545 | 14.68 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr12_+_57623477 | 14.56 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr6_-_17706618 | 14.53 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
| chr5_+_172386419 | 14.23 |
ENST00000265100.2
ENST00000519239.1 |
RPL26L1
|
ribosomal protein L26-like 1 |
| chr2_-_175113301 | 14.19 |
ENST00000344357.5
ENST00000284719.3 |
OLA1
|
Obg-like ATPase 1 |
| chr6_-_153304148 | 14.13 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chr1_-_86174065 | 14.10 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr9_+_131445928 | 14.09 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chr17_-_47492236 | 14.00 |
ENST00000434917.2
ENST00000300408.3 ENST00000511832.1 ENST00000419140.2 |
PHB
|
prohibitin |
| chr12_-_46766577 | 13.90 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr2_-_47142884 | 13.89 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chrX_-_16887963 | 13.71 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr1_+_214776516 | 13.52 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr5_+_110074685 | 13.37 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr2_+_187350973 | 13.35 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr15_-_49447835 | 13.24 |
ENST00000388901.5
ENST00000299259.6 |
COPS2
|
COP9 signalosome subunit 2 |
| chr6_-_114292449 | 13.16 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr22_-_42343117 | 13.05 |
ENST00000407253.3
ENST00000215980.5 |
CENPM
|
centromere protein M |
| chr1_+_38478432 | 12.98 |
ENST00000537711.1
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr12_+_109535923 | 12.74 |
ENST00000336865.2
|
UNG
|
uracil-DNA glycosylase |
| chr2_+_201171064 | 12.46 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_+_118572226 | 12.41 |
ENST00000263239.2
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr13_+_50656307 | 12.19 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr11_-_118972575 | 11.97 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr21_-_27542972 | 11.90 |
ENST00000346798.3
ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP
|
amyloid beta (A4) precursor protein |
| chr5_+_95066823 | 11.87 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr10_-_120938303 | 11.81 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr6_-_114292284 | 11.80 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr1_+_24018269 | 11.76 |
ENST00000374550.3
|
RPL11
|
ribosomal protein L11 |
| chr8_-_62627057 | 11.70 |
ENST00000519234.1
ENST00000379449.6 ENST00000379454.4 ENST00000518068.1 ENST00000517856.1 ENST00000356457.5 |
ASPH
|
aspartate beta-hydroxylase |
| chr1_+_111991474 | 11.59 |
ENST00000369722.3
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr11_+_69455855 | 11.50 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr2_-_44223138 | 11.37 |
ENST00000260665.7
|
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr14_+_64854958 | 11.16 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr15_-_101835414 | 11.04 |
ENST00000254193.6
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr10_-_71993176 | 10.99 |
ENST00000373232.3
|
PPA1
|
pyrophosphatase (inorganic) 1 |
| chr15_+_96875657 | 10.83 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr8_+_109455845 | 10.70 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr1_-_38061522 | 10.61 |
ENST00000373062.3
|
GNL2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr21_-_46237883 | 10.59 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr11_+_65769946 | 10.48 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr2_-_179315490 | 10.41 |
ENST00000487082.1
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr16_+_53088885 | 10.34 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_-_34637718 | 10.20 |
ENST00000378892.1
ENST00000277010.4 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr6_+_44214824 | 10.20 |
ENST00000371646.5
ENST00000353801.3 |
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr1_+_111992064 | 10.11 |
ENST00000483994.1
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr19_+_21688366 | 10.03 |
ENST00000358491.4
ENST00000597078.1 |
ZNF429
|
zinc finger protein 429 |
| chr5_-_176900610 | 10.01 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr6_+_44187334 | 10.01 |
ENST00000313248.7
ENST00000427851.2 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr3_-_183967296 | 9.97 |
ENST00000455059.1
ENST00000445626.2 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr2_+_64069459 | 9.93 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr16_+_14980632 | 9.83 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr11_+_71938925 | 9.82 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr4_+_57333756 | 9.81 |
ENST00000510663.1
ENST00000504757.1 |
SRP72
|
signal recognition particle 72kDa |
| chr1_-_43833628 | 9.76 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr4_+_41258786 | 9.73 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr12_-_49318715 | 9.62 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr11_-_47664072 | 9.58 |
ENST00000542981.1
ENST00000530428.1 ENST00000302503.3 |
MTCH2
|
mitochondrial carrier 2 |
| chr14_-_58894332 | 9.44 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr9_-_2844058 | 9.44 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr20_+_31407692 | 9.28 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr4_+_17579110 | 9.22 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr17_+_65713925 | 9.16 |
ENST00000253247.4
|
NOL11
|
nucleolar protein 11 |
| chr6_-_32811771 | 9.16 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr13_+_27998681 | 9.13 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr4_+_17578815 | 8.99 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr11_-_122932730 | 8.89 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr1_-_9129895 | 8.81 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr4_-_103266626 | 8.77 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr20_-_49547910 | 8.76 |
ENST00000396032.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr9_-_130890662 | 8.75 |
ENST00000277462.5
ENST00000338961.6 |
PTGES2
|
prostaglandin E synthase 2 |
| chr6_+_44187242 | 8.64 |
ENST00000393844.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr1_+_26496362 | 8.64 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr7_+_100464760 | 8.64 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr1_-_36107445 | 8.50 |
ENST00000373237.3
|
PSMB2
|
proteasome (prosome, macropain) subunit, beta type, 2 |
| chr6_-_153304697 | 8.46 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr5_+_172386517 | 8.45 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26-like 1 |
| chr2_+_68384976 | 8.43 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr6_+_44191507 | 8.39 |
ENST00000371724.1
ENST00000371713.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr1_+_24019099 | 8.38 |
ENST00000443624.1
ENST00000458455.1 |
RPL11
|
ribosomal protein L11 |
| chr22_+_18593446 | 8.28 |
ENST00000316027.6
|
TUBA8
|
tubulin, alpha 8 |
| chr3_-_50329990 | 8.18 |
ENST00000417626.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr3_+_124449213 | 8.17 |
ENST00000232607.2
ENST00000536109.1 ENST00000538242.1 ENST00000413078.2 |
UMPS
|
uridine monophosphate synthetase |
| chr1_-_40041925 | 8.08 |
ENST00000372862.3
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr6_-_160210604 | 8.07 |
ENST00000420894.2
ENST00000539756.1 ENST00000544255.1 |
TCP1
|
t-complex 1 |
| chr8_-_103424986 | 7.94 |
ENST00000521922.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr2_-_216257849 | 7.89 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr1_-_235292250 | 7.78 |
ENST00000366607.4
|
TOMM20
|
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chrX_+_51636629 | 7.67 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr1_+_32479430 | 7.64 |
ENST00000327300.7
ENST00000492989.1 |
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr22_+_20105259 | 7.63 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr1_-_40042073 | 7.62 |
ENST00000372858.3
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr16_+_771663 | 7.61 |
ENST00000568916.1
|
FAM173A
|
family with sequence similarity 173, member A |
| chr19_+_36036477 | 7.58 |
ENST00000222284.5
ENST00000392204.2 |
TMEM147
|
transmembrane protein 147 |
| chr13_-_95953589 | 7.56 |
ENST00000538287.1
ENST00000376887.4 ENST00000412704.1 ENST00000536256.1 ENST00000431522.1 |
ABCC4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr19_+_36036583 | 7.54 |
ENST00000392205.1
|
TMEM147
|
transmembrane protein 147 |
| chr6_-_97345689 | 7.51 |
ENST00000316149.7
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr10_-_16859361 | 7.49 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr8_-_117768023 | 7.48 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr2_-_179315453 | 7.45 |
ENST00000432031.2
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr5_+_145826867 | 7.45 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr6_-_47277634 | 7.35 |
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr11_-_76091986 | 7.18 |
ENST00000260045.3
|
PRKRIR
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) |
| chr12_+_6309963 | 7.10 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr3_+_133293278 | 6.96 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr9_-_6015607 | 6.96 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr16_-_69364467 | 6.95 |
ENST00000288022.1
|
PDF
|
peptide deformylase (mitochondrial) |
| chr3_+_133292574 | 6.92 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr6_+_44191290 | 6.90 |
ENST00000371755.3
ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr21_-_46238034 | 6.88 |
ENST00000332859.6
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr10_-_16859442 | 6.86 |
ENST00000602389.1
ENST00000345264.5 |
RSU1
|
Ras suppressor protein 1 |
| chr5_+_102455968 | 6.83 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr3_-_160283348 | 6.83 |
ENST00000334256.4
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3) |
| chr17_+_7476136 | 6.74 |
ENST00000582169.1
ENST00000578754.1 ENST00000578495.1 ENST00000293831.8 ENST00000380512.5 ENST00000585024.1 ENST00000583802.1 ENST00000577269.1 ENST00000584784.1 ENST00000582746.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr5_+_70883117 | 6.71 |
ENST00000340941.6
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr6_+_149539767 | 6.68 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr2_+_32853093 | 6.66 |
ENST00000448773.1
ENST00000317907.4 |
TTC27
|
tetratricopeptide repeat domain 27 |
| chr12_+_108079509 | 6.63 |
ENST00000412830.3
ENST00000547995.1 |
PWP1
|
PWP1 homolog (S. cerevisiae) |
| chr17_-_7590745 | 6.54 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr1_+_110163709 | 6.52 |
ENST00000369840.2
ENST00000527846.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr3_-_98312548 | 6.49 |
ENST00000264193.2
|
CPOX
|
coproporphyrinogen oxidase |
| chr8_-_103424916 | 6.40 |
ENST00000220959.4
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr7_+_26191809 | 6.38 |
ENST00000056233.3
|
NFE2L3
|
nuclear factor, erythroid 2-like 3 |
| chr11_-_94227029 | 6.35 |
ENST00000323977.3
ENST00000536754.1 ENST00000323929.3 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr19_+_50180409 | 6.25 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr20_+_48552908 | 6.24 |
ENST00000244061.2
|
RNF114
|
ring finger protein 114 |
| chr5_-_115177247 | 6.24 |
ENST00000500945.2
|
ATG12
|
autophagy related 12 |
| chr1_-_154909329 | 6.20 |
ENST00000368467.3
|
PMVK
|
phosphomevalonate kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of CLOCK
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.6 | 68.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 11.8 | 11.8 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 11.6 | 81.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 10.8 | 43.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 10.4 | 52.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 9.3 | 46.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 8.9 | 70.8 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) positive regulation of exit from mitosis(GO:0031536) |
| 7.5 | 22.6 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 7.5 | 44.7 | GO:2000323 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 6.8 | 33.9 | GO:0015862 | uridine transport(GO:0015862) |
| 6.4 | 31.8 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 6.3 | 50.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 5.9 | 53.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 5.6 | 11.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 5.5 | 82.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 5.2 | 26.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 5.0 | 20.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 5.0 | 25.0 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 4.8 | 28.8 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 4.6 | 13.9 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 4.6 | 18.3 | GO:1901165 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of trophoblast cell migration(GO:1901165) |
| 4.5 | 13.4 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 3.9 | 15.6 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 3.9 | 11.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 3.8 | 15.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 3.7 | 11.0 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 3.4 | 16.8 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 3.2 | 9.7 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.1 | 50.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 3.1 | 12.5 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 3.1 | 9.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 2.7 | 13.7 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 2.7 | 8.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 2.7 | 10.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 2.7 | 18.6 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 2.6 | 7.9 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 2.6 | 7.8 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 2.4 | 11.9 | GO:0071873 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 2.4 | 14.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 2.3 | 13.9 | GO:0032328 | alanine transport(GO:0032328) |
| 2.3 | 27.5 | GO:0015074 | DNA integration(GO:0015074) |
| 2.2 | 29.0 | GO:0045008 | depyrimidination(GO:0045008) |
| 2.2 | 6.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 2.1 | 6.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 2.1 | 6.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 2.0 | 9.9 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 2.0 | 11.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.9 | 7.6 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 1.8 | 5.4 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 1.8 | 72.0 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 1.7 | 8.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 1.7 | 8.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 1.7 | 10.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 1.7 | 5.0 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 1.7 | 11.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.7 | 16.5 | GO:0006983 | ER overload response(GO:0006983) |
| 1.6 | 9.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 1.6 | 14.5 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 1.6 | 4.8 | GO:0050894 | determination of affect(GO:0050894) |
| 1.6 | 4.8 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.6 | 4.7 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 1.6 | 15.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 1.5 | 12.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 1.5 | 3.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 1.5 | 10.6 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 1.5 | 4.4 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.3 | 18.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 1.3 | 9.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 1.3 | 8.8 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 1.2 | 18.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 1.2 | 6.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.2 | 38.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.2 | 10.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 1.2 | 11.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 1.2 | 7.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.1 | 11.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 1.1 | 3.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.0 | 13.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 1.0 | 13.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 1.0 | 17.9 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 1.0 | 5.7 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.9 | 8.5 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.9 | 2.8 | GO:0048627 | myoblast development(GO:0048627) |
| 0.9 | 5.4 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.9 | 8.9 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.9 | 15.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.9 | 13.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.9 | 14.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.8 | 5.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.8 | 8.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.8 | 4.9 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.8 | 1.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.8 | 16.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.7 | 2.2 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.7 | 19.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.7 | 7.1 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.7 | 2.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.7 | 8.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.7 | 2.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.7 | 32.6 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.6 | 6.5 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.6 | 14.1 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.6 | 10.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.6 | 29.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.6 | 6.9 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.6 | 3.9 | GO:0090032 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.6 | 5.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.5 | 19.2 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.5 | 7.6 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.5 | 3.7 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.5 | 22.2 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.5 | 4.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 8.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 13.0 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.5 | 4.4 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.5 | 6.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.5 | 27.5 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.5 | 6.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.5 | 9.1 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.4 | 13.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.4 | 1.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.4 | 38.6 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.4 | 2.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 11.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 | 1.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.4 | 1.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.4 | 5.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.4 | 25.8 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.4 | 1.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.4 | 2.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 3.5 | GO:0006188 | IMP biosynthetic process(GO:0006188) |
| 0.3 | 2.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.3 | 2.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 6.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.3 | 4.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 3.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 5.1 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 1.9 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) negative regulation of lamellipodium organization(GO:1902744) |
| 0.2 | 4.9 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.2 | 22.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 3.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 9.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 12.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 9.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 1.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 37.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.2 | 1.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 6.7 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.2 | 4.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 10.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 11.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.4 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 3.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 2.6 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 2.7 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 4.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 21.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 4.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 1.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 5.6 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.1 | 1.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 1.3 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 8.5 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 1.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 2.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 10.6 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 13.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 14.2 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.1 | 1.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.8 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 2.6 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 5.5 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 3.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.4 | GO:0035150 | regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.1 | 3.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 1.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.3 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.3 | GO:0051252 | regulation of RNA metabolic process(GO:0051252) |
| 0.0 | 1.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 1.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 5.5 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 1.7 | GO:0006401 | RNA catabolic process(GO:0006401) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 48.6 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 10.1 | 81.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 8.7 | 26.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 8.7 | 52.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 4.5 | 31.8 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 3.9 | 15.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 3.8 | 15.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 3.7 | 14.7 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 3.5 | 21.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 3.1 | 9.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 2.4 | 70.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 2.1 | 10.7 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 2.1 | 38.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 2.1 | 29.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 2.0 | 16.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.9 | 11.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 1.9 | 11.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.9 | 33.9 | GO:0034709 | methylosome(GO:0034709) |
| 1.8 | 14.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 1.7 | 8.6 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.7 | 5.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 1.5 | 9.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.5 | 18.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 1.5 | 12.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 1.5 | 38.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.5 | 14.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.3 | 58.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 1.3 | 17.9 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 1.2 | 31.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 1.2 | 9.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 1.2 | 86.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 1.2 | 10.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.1 | 9.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.1 | 7.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 1.0 | 6.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 1.0 | 86.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 1.0 | 50.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.9 | 8.5 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.9 | 3.8 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.9 | 11.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.9 | 6.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.8 | 7.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.8 | 7.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.7 | 11.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.7 | 8.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.7 | 2.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.6 | 7.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 16.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.5 | 7.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.5 | 3.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 2.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.4 | 4.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.4 | 4.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.4 | 5.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 42.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 18.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 5.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.3 | 33.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.3 | 36.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.3 | 77.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 2.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 33.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.3 | 13.9 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 4.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 8.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.3 | 3.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 18.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 27.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 3.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 2.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 28.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.2 | 9.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 3.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 7.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 2.9 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.2 | 17.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 13.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 2.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 17.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 24.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 8.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 4.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 5.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 21.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 30.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 114.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 28.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 9.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 33.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 3.3 | GO:0044391 | ribosomal subunit(GO:0044391) |
| 0.1 | 1.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 4.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 11.2 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 0.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 2.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.6 | 81.1 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 11.2 | 44.7 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 10.8 | 75.8 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 7.9 | 47.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 7.3 | 21.9 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 7.3 | 58.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 7.3 | 21.8 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 6.5 | 26.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 6.5 | 19.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 6.1 | 18.3 | GO:0030984 | kininogen binding(GO:0030984) |
| 5.2 | 15.6 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 4.8 | 28.8 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 4.3 | 42.7 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 4.0 | 20.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 4.0 | 12.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 3.5 | 27.7 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 3.3 | 10.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 3.3 | 9.9 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 3.3 | 9.8 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 3.0 | 21.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.9 | 31.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 2.9 | 8.6 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 2.7 | 11.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 2.4 | 9.7 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.2 | 8.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 2.2 | 10.8 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 2.1 | 10.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 2.1 | 23.3 | GO:0031386 | protein tag(GO:0031386) |
| 2.0 | 46.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 2.0 | 17.9 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.9 | 72.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 1.9 | 7.6 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 1.8 | 5.4 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 1.8 | 25.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 1.6 | 67.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 1.6 | 33.9 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 1.6 | 4.8 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 1.6 | 11.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 1.6 | 71.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 1.5 | 10.6 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 1.5 | 9.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 1.4 | 38.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.4 | 4.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 1.3 | 12.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.2 | 18.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 1.2 | 9.8 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 1.2 | 11.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.2 | 55.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 1.2 | 28.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 1.2 | 7.0 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.1 | 5.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.0 | 3.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.9 | 14.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.9 | 5.4 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.9 | 4.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.9 | 4.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.8 | 13.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.8 | 15.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.8 | 4.9 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.8 | 36.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.8 | 7.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.8 | 83.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.7 | 13.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.7 | 1.4 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.7 | 2.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.7 | 19.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.7 | 2.7 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.7 | 2.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.6 | 18.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.6 | 8.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.6 | 13.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.6 | 12.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.6 | 17.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.6 | 4.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.6 | 2.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 10.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.5 | 6.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.5 | 2.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.5 | 1.6 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.5 | 20.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.5 | 5.7 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.5 | 5.0 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.5 | 3.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.4 | 9.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 8.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.4 | 4.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) O-acetyltransferase activity(GO:0016413) |
| 0.4 | 10.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 7.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 6.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 9.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.4 | 3.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 7.0 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 3.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.4 | 10.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.4 | 6.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 16.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.3 | 3.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 9.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 2.6 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.3 | 1.9 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 18.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.3 | 10.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 10.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.3 | 3.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 8.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 7.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.3 | 18.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 16.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 112.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 8.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.2 | 3.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 35.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 7.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 3.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 2.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 10.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 2.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 3.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.2 | 5.6 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.2 | 6.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 13.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 14.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 4.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.8 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 6.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 6.2 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 5.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 2.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 25.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 4.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 6.1 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 7.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 10.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 14.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 5.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.1 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 2.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 16.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 74.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 2.3 | 55.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.7 | 99.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.6 | 14.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 41.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.5 | 16.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 41.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.4 | 11.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 30.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.4 | 7.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.4 | 9.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 13.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.3 | 31.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.3 | 32.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.3 | 16.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 14.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.3 | 11.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 9.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 14.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 9.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 10.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 9.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 26.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 7.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 3.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 9.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 4.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 3.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 2.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 93.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 3.2 | 28.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 2.5 | 70.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 1.7 | 47.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 1.7 | 38.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 1.5 | 27.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.3 | 33.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 1.3 | 40.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.3 | 15.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.1 | 25.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.1 | 61.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 1.1 | 6.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 1.0 | 25.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 1.0 | 15.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 1.0 | 27.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.9 | 61.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.9 | 18.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 6.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.7 | 5.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.7 | 31.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.6 | 29.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.6 | 10.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.5 | 7.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.5 | 31.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.5 | 4.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.5 | 14.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 18.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.5 | 16.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 6.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 3.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.4 | 10.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 9.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 7.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.4 | 23.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.4 | 11.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.4 | 7.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 52.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 2.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 13.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.3 | 6.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 9.8 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.3 | 22.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 13.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 17.7 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.3 | 9.9 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.2 | 5.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 6.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 6.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 6.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 5.7 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 10.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 6.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 3.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 6.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 4.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 7.6 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 2.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.5 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 6.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 5.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 10.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 4.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 8.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 3.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 4.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |