Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
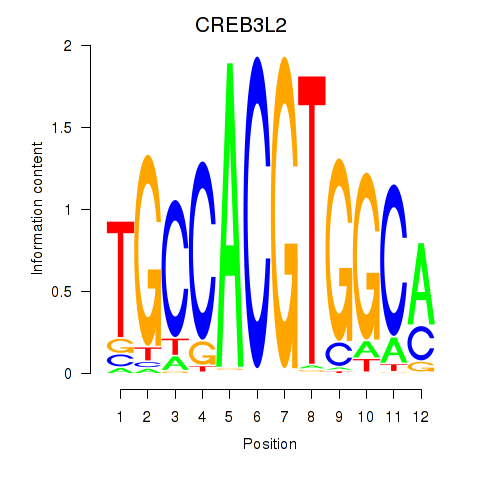
Results for CREB3L2
Z-value: 1.31
Transcription factors associated with CREB3L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L2
|
ENSG00000182158.10 | cAMP responsive element binding protein 3 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L2 | hg19_v2_chr7_-_137686791_137686821 | 0.60 | 5.1e-23 | Click! |
Activity profile of CREB3L2 motif
Sorted Z-values of CREB3L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_6523688 | 23.92 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr8_+_22224760 | 22.04 |
ENST00000359741.5
ENST00000520644.1 ENST00000240095.6 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr2_+_74425689 | 21.63 |
ENST00000394053.2
ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr6_-_7911042 | 20.27 |
ENST00000379757.4
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr12_-_56123444 | 18.39 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr12_-_49318715 | 18.23 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr12_-_56122761 | 17.70 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr1_-_11120057 | 17.17 |
ENST00000376957.2
|
SRM
|
spermidine synthase |
| chr19_-_4670345 | 17.01 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr21_-_18985158 | 16.53 |
ENST00000339775.6
|
BTG3
|
BTG family, member 3 |
| chr21_-_18985230 | 16.21 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr7_-_6523755 | 16.15 |
ENST00000436575.1
ENST00000258739.4 |
DAGLB
KDELR2
|
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr2_-_10588630 | 15.21 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr2_-_69614373 | 15.03 |
ENST00000361060.5
ENST00000357308.4 |
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr21_-_40720974 | 13.80 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr17_+_49243792 | 13.69 |
ENST00000393183.3
ENST00000393190.1 |
NME1-NME2
|
NME1-NME2 readthrough |
| chr21_-_40720995 | 13.67 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chrX_+_48433326 | 13.62 |
ENST00000376755.1
|
RBM3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr5_+_34656569 | 13.45 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr8_+_82192501 | 13.35 |
ENST00000297258.6
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated) |
| chr7_-_121036337 | 13.11 |
ENST00000426156.1
ENST00000359943.3 ENST00000412653.1 |
FAM3C
|
family with sequence similarity 3, member C |
| chr11_+_32112431 | 13.01 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr1_+_46016703 | 12.29 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr5_+_34656331 | 12.28 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr3_-_57583052 | 12.06 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr6_-_32811771 | 11.74 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr12_+_57624085 | 11.54 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr3_+_122785895 | 11.52 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr8_-_144679296 | 11.31 |
ENST00000317198.6
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chrX_+_48432892 | 11.25 |
ENST00000376759.3
ENST00000430348.2 |
RBM3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr6_+_13615554 | 11.03 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chr3_-_57583185 | 10.74 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr2_+_216176540 | 10.68 |
ENST00000236959.9
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr5_+_112196919 | 10.59 |
ENST00000505459.1
ENST00000282999.3 ENST00000515463.1 |
SRP19
|
signal recognition particle 19kDa |
| chr3_-_57583130 | 10.51 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr2_+_113403434 | 10.41 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr7_+_100464760 | 10.35 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr12_+_57624119 | 9.87 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr2_+_216176761 | 9.73 |
ENST00000540518.1
ENST00000435675.1 |
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr10_-_71993176 | 9.48 |
ENST00000373232.3
|
PPA1
|
pyrophosphatase (inorganic) 1 |
| chr5_+_65440032 | 9.44 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr2_+_198365095 | 9.26 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr2_-_47143160 | 9.23 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr17_+_49243639 | 9.20 |
ENST00000512737.1
ENST00000503064.1 |
NME1-NME2
|
NME1-NME2 readthrough |
| chr10_+_89419370 | 9.09 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr1_+_42922173 | 8.95 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr12_+_57623869 | 8.95 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr3_+_148709128 | 8.83 |
ENST00000345003.4
ENST00000296048.6 ENST00000483267.1 |
GYG1
|
glycogenin 1 |
| chr5_+_122110691 | 8.66 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr1_+_42921761 | 8.41 |
ENST00000372562.1
|
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr5_-_143550159 | 8.40 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr2_-_47142884 | 8.28 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr3_+_158519654 | 8.11 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr8_-_144679532 | 7.98 |
ENST00000534380.1
ENST00000533494.1 ENST00000531218.1 ENST00000526340.1 ENST00000533204.1 ENST00000532400.1 ENST00000529516.1 ENST00000534377.1 ENST00000531621.1 ENST00000530191.1 ENST00000524900.1 ENST00000526838.1 ENST00000531931.1 ENST00000534475.1 ENST00000442189.2 ENST00000524624.1 ENST00000532596.1 ENST00000529832.1 ENST00000530306.1 ENST00000530545.1 ENST00000525261.1 ENST00000534804.1 ENST00000528303.1 ENST00000528610.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr2_-_27545921 | 7.92 |
ENST00000402310.1
ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr20_+_44520009 | 7.88 |
ENST00000607482.1
ENST00000372459.2 |
CTSA
|
cathepsin A |
| chr1_+_180123969 | 7.78 |
ENST00000367602.3
ENST00000367600.5 |
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr2_-_65357225 | 7.61 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr20_+_44519948 | 7.60 |
ENST00000354880.5
ENST00000191018.5 |
CTSA
|
cathepsin A |
| chr22_-_43411106 | 7.60 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr2_+_172544294 | 7.58 |
ENST00000358002.6
ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr12_+_57623477 | 7.50 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr18_+_158513 | 7.30 |
ENST00000400266.3
ENST00000580410.1 ENST00000383589.2 ENST00000261601.7 |
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr2_+_46926048 | 7.17 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr10_-_120840309 | 7.16 |
ENST00000369144.3
|
EIF3A
|
eukaryotic translation initiation factor 3, subunit A |
| chr17_-_39968855 | 7.14 |
ENST00000355468.3
ENST00000590496.1 |
LEPREL4
|
leprecan-like 4 |
| chr17_+_39969183 | 7.10 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr10_-_126849588 | 7.04 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr3_+_148709310 | 7.04 |
ENST00000484197.1
ENST00000492285.2 ENST00000461191.1 |
GYG1
|
glycogenin 1 |
| chr19_+_47634039 | 7.02 |
ENST00000597808.1
ENST00000413379.3 ENST00000600706.1 ENST00000540850.1 ENST00000598840.1 ENST00000600753.1 ENST00000270225.7 ENST00000392776.3 |
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr2_+_201676256 | 6.98 |
ENST00000452206.1
ENST00000410110.2 ENST00000409600.1 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr4_-_83812402 | 6.92 |
ENST00000395310.2
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr19_+_36705504 | 6.85 |
ENST00000456324.1
|
ZNF146
|
zinc finger protein 146 |
| chr19_-_42759300 | 6.65 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr2_+_201676908 | 6.64 |
ENST00000409226.1
ENST00000452790.2 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chrX_+_131157322 | 6.58 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr2_+_171785824 | 6.53 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr9_+_117350009 | 6.50 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chr4_-_83812248 | 6.49 |
ENST00000514326.1
ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr5_-_143550241 | 6.42 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr10_+_103892787 | 6.39 |
ENST00000278070.2
ENST00000413464.2 |
PPRC1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr10_+_112257596 | 6.29 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chrX_+_131157290 | 6.25 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr10_-_97416400 | 6.24 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr2_-_70520539 | 6.08 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr11_-_62341445 | 6.06 |
ENST00000329251.4
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma |
| chr12_-_76953573 | 5.99 |
ENST00000549646.1
ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr22_-_43253189 | 5.89 |
ENST00000437119.2
ENST00000429508.2 ENST00000454099.1 ENST00000263245.5 |
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr15_-_72668805 | 5.86 |
ENST00000268097.5
|
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr10_-_71930222 | 5.75 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr11_-_118972575 | 5.71 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr6_-_43197189 | 5.65 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr16_-_30441293 | 5.65 |
ENST00000565758.1
ENST00000567983.1 ENST00000319285.4 |
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr2_+_207024306 | 5.59 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr10_+_70715884 | 5.56 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr16_+_88923494 | 5.43 |
ENST00000567895.1
ENST00000301021.3 ENST00000565504.1 ENST00000567312.1 ENST00000568583.1 ENST00000561840.1 |
TRAPPC2L
|
trafficking protein particle complex 2-like |
| chr17_+_8191815 | 5.37 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr18_+_9913977 | 5.36 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr19_+_36706024 | 5.32 |
ENST00000443387.2
|
ZNF146
|
zinc finger protein 146 |
| chr2_+_172544182 | 5.04 |
ENST00000409197.1
ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr6_+_31802685 | 4.94 |
ENST00000375639.2
ENST00000375638.3 ENST00000375635.2 ENST00000375642.2 ENST00000395789.1 |
C6orf48
|
chromosome 6 open reading frame 48 |
| chr1_-_17380630 | 4.93 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr12_+_131356582 | 4.87 |
ENST00000448750.3
ENST00000541630.1 ENST00000392369.2 ENST00000254675.3 ENST00000535090.1 ENST00000392367.3 |
RAN
|
RAN, member RAS oncogene family |
| chr11_-_61560053 | 4.78 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr6_+_32811885 | 4.72 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chrX_-_102941596 | 4.60 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr7_-_134143841 | 4.59 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr6_+_31802364 | 4.53 |
ENST00000375640.3
ENST00000375641.2 |
C6orf48
|
chromosome 6 open reading frame 48 |
| chr2_+_27440229 | 4.49 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr7_+_16793160 | 4.30 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr1_+_32479430 | 4.29 |
ENST00000327300.7
ENST00000492989.1 |
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chrX_+_105937068 | 4.23 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr6_-_144329531 | 4.15 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr19_-_47290535 | 4.13 |
ENST00000412532.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr15_+_89182178 | 4.08 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr6_+_151561506 | 3.92 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr1_+_116915855 | 3.92 |
ENST00000295598.5
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr6_-_32140886 | 3.86 |
ENST00000395496.1
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr2_+_131100710 | 3.76 |
ENST00000452955.1
|
IMP4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr11_-_6502534 | 3.74 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr19_+_34663397 | 3.63 |
ENST00000540746.2
ENST00000544216.3 ENST00000433627.5 |
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae) |
| chr9_+_706842 | 3.54 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr1_+_44444865 | 3.50 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr9_-_125667494 | 3.42 |
ENST00000335387.5
ENST00000357244.2 ENST00000373665.2 |
RC3H2
|
ring finger and CCCH-type domains 2 |
| chr6_-_28891709 | 3.40 |
ENST00000377194.3
ENST00000377199.3 |
TRIM27
|
tripartite motif containing 27 |
| chr2_+_217498105 | 3.33 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr22_-_18257178 | 3.29 |
ENST00000342111.5
|
BID
|
BH3 interacting domain death agonist |
| chr2_+_201936458 | 3.27 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr8_+_56014949 | 3.10 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr15_+_89181974 | 3.05 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_+_46926326 | 2.99 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr3_+_133292759 | 2.97 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr1_-_153940097 | 2.93 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr2_+_172543919 | 2.92 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr5_-_150537279 | 2.90 |
ENST00000517486.1
ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6
|
annexin A6 |
| chr2_+_169312725 | 2.87 |
ENST00000392687.4
|
CERS6
|
ceramide synthase 6 |
| chr2_+_172544011 | 2.85 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr1_-_32801825 | 2.82 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr3_+_12525931 | 2.81 |
ENST00000446004.1
ENST00000314571.7 ENST00000454502.2 ENST00000383797.5 ENST00000402228.3 ENST00000284995.6 ENST00000444864.1 |
TSEN2
|
TSEN2 tRNA splicing endonuclease subunit |
| chr11_+_111957497 | 2.77 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr22_-_18256742 | 2.75 |
ENST00000317361.7
|
BID
|
BH3 interacting domain death agonist |
| chr20_-_57607347 | 2.74 |
ENST00000395663.1
ENST00000395659.1 ENST00000243997.3 |
ATP5E
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr2_-_120980939 | 2.72 |
ENST00000426077.2
|
TMEM185B
|
transmembrane protein 185B |
| chr2_+_172543967 | 2.70 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr7_+_150929550 | 2.67 |
ENST00000482173.1
ENST00000495645.1 ENST00000035307.2 |
CHPF2
|
chondroitin polymerizing factor 2 |
| chr18_+_9708162 | 2.66 |
ENST00000578921.1
|
RAB31
|
RAB31, member RAS oncogene family |
| chr12_+_6833437 | 2.44 |
ENST00000534947.1
ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr22_+_38864041 | 2.42 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr2_-_27341765 | 2.25 |
ENST00000405600.1
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr1_-_45956800 | 2.21 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr1_-_1167411 | 2.19 |
ENST00000263741.7
|
SDF4
|
stromal cell derived factor 4 |
| chr3_+_45636219 | 2.14 |
ENST00000273317.4
|
LIMD1
|
LIM domains containing 1 |
| chr1_+_231376941 | 2.12 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr12_+_6833237 | 2.12 |
ENST00000229251.3
ENST00000539735.1 ENST00000538410.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr22_-_18257249 | 2.10 |
ENST00000399765.1
ENST00000399767.1 ENST00000399774.3 |
BID
|
BH3 interacting domain death agonist |
| chr21_+_37442239 | 2.05 |
ENST00000530908.1
ENST00000290349.6 ENST00000439427.2 ENST00000399191.3 |
CBR1
|
carbonyl reductase 1 |
| chr8_-_99129338 | 1.96 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr19_+_34663551 | 1.87 |
ENST00000586157.1
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae) |
| chr17_+_65821780 | 1.81 |
ENST00000321892.4
ENST00000335221.5 ENST00000306378.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr14_-_106174960 | 1.79 |
ENST00000390547.2
|
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr3_-_155572164 | 1.72 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr11_-_111957451 | 1.71 |
ENST00000504148.2
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr12_-_93836028 | 1.68 |
ENST00000318066.2
|
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr5_+_43603229 | 1.68 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr12_-_93835665 | 1.65 |
ENST00000552442.1
ENST00000550657.1 |
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr8_+_1772132 | 1.64 |
ENST00000349830.3
ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr2_+_169312350 | 1.59 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chr15_+_75315896 | 1.46 |
ENST00000342932.3
ENST00000564923.1 ENST00000569562.1 ENST00000568649.1 |
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr20_+_25228669 | 1.39 |
ENST00000216962.4
|
PYGB
|
phosphorylase, glycogen; brain |
| chr14_+_101193164 | 1.38 |
ENST00000341267.4
|
DLK1
|
delta-like 1 homolog (Drosophila) |
| chr6_+_151561085 | 1.36 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr3_-_123603137 | 1.31 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr20_-_33413416 | 1.31 |
ENST00000359003.2
|
NCOA6
|
nuclear receptor coactivator 6 |
| chr15_-_72668185 | 1.30 |
ENST00000457859.2
ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr11_-_6502580 | 1.28 |
ENST00000423813.2
ENST00000396777.3 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr4_-_186347099 | 1.24 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr12_+_67663056 | 1.24 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr12_-_104531945 | 1.23 |
ENST00000551446.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr9_-_125667618 | 1.15 |
ENST00000423239.2
|
RC3H2
|
ring finger and CCCH-type domains 2 |
| chr2_-_27341966 | 1.00 |
ENST00000402394.1
ENST00000402550.1 ENST00000260595.5 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr19_-_44143939 | 0.99 |
ENST00000222374.2
|
CADM4
|
cell adhesion molecule 4 |
| chr20_+_61584026 | 0.92 |
ENST00000370351.4
ENST00000370349.3 |
SLC17A9
|
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
| chr12_-_6716534 | 0.91 |
ENST00000544484.1
ENST00000309577.6 ENST00000357008.2 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr2_-_27341872 | 0.85 |
ENST00000312734.4
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr15_+_58724184 | 0.85 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr14_-_106054659 | 0.84 |
ENST00000390539.2
|
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr22_-_29663954 | 0.79 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chr1_-_113498943 | 0.75 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr12_-_6716569 | 0.75 |
ENST00000544040.1
ENST00000545942.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr13_+_98794810 | 0.74 |
ENST00000595437.1
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr9_+_130565487 | 0.70 |
ENST00000373225.3
ENST00000431857.1 |
FPGS
|
folylpolyglutamate synthase |
| chr17_+_80477571 | 0.69 |
ENST00000335255.5
|
FOXK2
|
forkhead box K2 |
| chr7_-_100808843 | 0.68 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr12_+_121416437 | 0.62 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr21_+_44394742 | 0.56 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr19_+_10812108 | 0.48 |
ENST00000250237.5
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase 1 |
| chr5_-_114880533 | 0.43 |
ENST00000274457.3
|
FEM1C
|
fem-1 homolog c (C. elegans) |
| chr19_+_49458107 | 0.41 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr17_-_2169425 | 0.39 |
ENST00000570606.1
ENST00000354901.4 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr20_-_17662878 | 0.37 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 37.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 5.1 | 20.4 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 4.1 | 12.3 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 3.8 | 15.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 3.8 | 41.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 3.5 | 10.6 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 3.2 | 9.5 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 3.0 | 33.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) apical protein localization(GO:0045176) |
| 2.8 | 36.1 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 2.5 | 17.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 2.4 | 7.2 | GO:0002188 | formation of cytoplasmic translation initiation complex(GO:0001732) translation reinitiation(GO:0002188) |
| 2.4 | 7.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 2.3 | 9.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 2.2 | 22.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 2.1 | 10.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 2.1 | 6.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.9 | 21.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 1.8 | 5.4 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 1.7 | 15.5 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 1.7 | 10.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 1.6 | 4.8 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 1.5 | 4.6 | GO:0006059 | hexitol metabolic process(GO:0006059) response to methylglyoxal(GO:0051595) |
| 1.5 | 4.6 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 1.5 | 4.5 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 1.3 | 5.4 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 1.3 | 26.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 1.2 | 3.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.1 | 5.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 1.1 | 14.8 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 1.1 | 5.7 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 1.1 | 15.0 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 1.1 | 4.3 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 1.0 | 10.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 1.0 | 6.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 1.0 | 4.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.9 | 27.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.9 | 2.8 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.9 | 2.8 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.9 | 18.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.9 | 6.0 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.8 | 8.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.8 | 7.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.8 | 7.6 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.7 | 2.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.7 | 4.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.7 | 3.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.6 | 8.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.6 | 3.9 | GO:0090032 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.5 | 7.9 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.5 | 4.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.5 | 9.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.5 | 39.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.4 | 16.1 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.4 | 5.5 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.4 | 0.4 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.4 | 18.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.4 | 6.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 4.9 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.4 | 6.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.4 | 2.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 5.7 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.4 | 5.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 1.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 1.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 4.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 2.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 14.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 1.7 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.3 | 3.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 7.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 3.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.2 | 5.0 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 30.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 2.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 0.8 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.2 | 6.1 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.2 | 0.6 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.2 | 10.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 0.6 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 16.5 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.2 | 1.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 4.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.1 | 1.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 5.6 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.1 | 3.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 16.9 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.1 | 2.8 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 2.9 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 1.7 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 30.8 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 4.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 2.2 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 4.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 2.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.4 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 7.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 2.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 2.7 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.1 | 3.0 | GO:0008283 | cell proliferation(GO:0008283) |
| 0.1 | 5.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 3.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.3 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 6.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.3 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.7 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 13.7 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.0 | 4.3 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 1.2 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 4.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 2.9 | GO:0006364 | rRNA processing(GO:0006364) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 37.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 3.0 | 36.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 2.5 | 24.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 2.2 | 15.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 2.1 | 10.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 2.1 | 16.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.9 | 7.7 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.8 | 7.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.4 | 8.7 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.3 | 10.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 1.2 | 33.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 1.1 | 4.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.1 | 3.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.9 | 2.6 | GO:0071751 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.8 | 7.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.8 | 6.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.8 | 3.8 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.7 | 13.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.7 | 21.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.7 | 2.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.6 | 34.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.5 | 7.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.5 | 5.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.4 | 4.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 2.7 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.3 | 4.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 44.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 6.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.3 | 1.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 26.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 2.2 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 7.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 3.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 2.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 7.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 3.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 24.9 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 17.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 8.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 2.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 8.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 4.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 4.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 14.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 7.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 5.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 5.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 10.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 11.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 7.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 1.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 4.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 3.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.7 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 6.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 9.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 9.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 12.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 15.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 5.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 30.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 1.6 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 10.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 2.6 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 40.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 7.2 | 21.6 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 6.8 | 20.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 5.7 | 17.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 5.4 | 37.8 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 4.0 | 15.9 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 3.1 | 22.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 3.0 | 9.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 2.6 | 10.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 2.6 | 7.7 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 2.4 | 7.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 2.4 | 9.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 2.1 | 6.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.9 | 7.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 1.9 | 15.5 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 1.9 | 5.7 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 1.9 | 16.9 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 1.8 | 5.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 1.7 | 8.7 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.6 | 30.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 1.6 | 6.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 1.5 | 4.5 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.3 | 33.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 1.3 | 3.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.2 | 10.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 1.2 | 7.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.1 | 30.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 1.1 | 6.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.0 | 7.0 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.0 | 19.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.9 | 2.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.9 | 7.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.9 | 2.7 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.9 | 25.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.8 | 17.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.8 | 5.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.7 | 4.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.7 | 5.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.7 | 15.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.6 | 29.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.6 | 5.7 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.6 | 4.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.6 | 1.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.6 | 11.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.6 | 2.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.5 | 16.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 4.9 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.5 | 10.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.5 | 2.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 6.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 2.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.4 | 7.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 1.4 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 5.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 16.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 3.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 3.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.3 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 8.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 17.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 3.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 2.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) O-acetyltransferase activity(GO:0016413) |
| 0.2 | 5.7 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.2 | 4.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 2.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 9.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 2.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 5.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 9.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 3.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 3.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 3.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 5.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 13.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 1.2 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 6.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 0.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 2.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 7.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 18.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 25.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 3.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 6.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.8 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 1.7 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 4.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 16.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 7.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 5.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 12.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 8.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 6.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 28.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 10.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 7.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 10.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 4.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 6.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 11.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 18.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 1.2 | 20.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.0 | 25.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.6 | 21.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.6 | 17.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.6 | 44.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.5 | 9.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.5 | 22.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.5 | 20.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 9.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.4 | 6.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.4 | 16.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 19.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.3 | 8.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 9.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 3.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.3 | 9.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 6.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 21.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 6.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 16.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 5.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 6.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 8.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 48.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.2 | 27.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 2.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 4.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 12.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 4.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 4.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 3.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 6.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 7.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 3.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 7.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |