Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
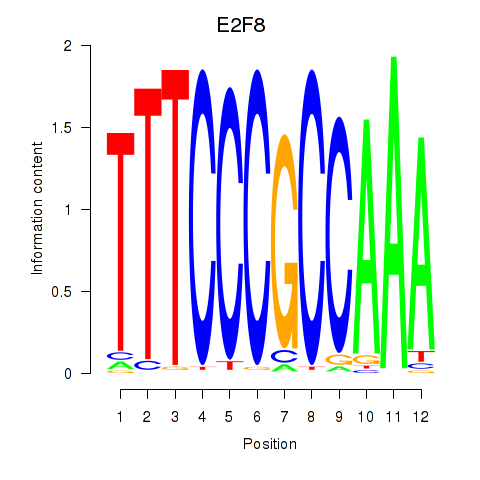
Results for E2F8
Z-value: 2.26
Transcription factors associated with E2F8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F8
|
ENSG00000129173.8 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F8 | hg19_v2_chr11_-_19263145_19263176 | 0.54 | 8.9e-18 | Click! |
Activity profile of E2F8 motif
Sorted Z-values of E2F8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_10262857 | 66.70 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr4_-_174254823 | 60.08 |
ENST00000438704.2
|
HMGB2
|
high mobility group box 2 |
| chr2_-_136633940 | 54.93 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr6_+_135502466 | 40.00 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_+_135502408 | 39.23 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_+_127317066 | 38.14 |
ENST00000265056.7
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr14_-_55658252 | 37.74 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr22_+_35796108 | 36.13 |
ENST00000382011.5
ENST00000416905.1 |
MCM5
|
minichromosome maintenance complex component 5 |
| chr10_-_17659234 | 36.06 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr8_+_128748308 | 34.77 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr1_-_159893507 | 34.66 |
ENST00000368096.1
|
TAGLN2
|
transgelin 2 |
| chr12_+_104682496 | 33.13 |
ENST00000378070.4
|
TXNRD1
|
thioredoxin reductase 1 |
| chr20_+_42295745 | 32.24 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr1_-_43638168 | 31.87 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr8_+_128748466 | 31.80 |
ENST00000524013.1
ENST00000520751.1 |
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr15_-_64673630 | 31.71 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr7_+_135242652 | 31.59 |
ENST00000285968.6
ENST00000440390.2 |
NUP205
|
nucleoporin 205kDa |
| chr9_+_131445928 | 31.44 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chr22_+_35796056 | 31.02 |
ENST00000216122.4
|
MCM5
|
minichromosome maintenance complex component 5 |
| chr12_-_57146095 | 30.65 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr14_-_55658323 | 30.14 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr15_+_64428529 | 29.42 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr14_+_51706886 | 28.87 |
ENST00000457354.2
|
TMX1
|
thioredoxin-related transmembrane protein 1 |
| chr2_-_152146385 | 28.00 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr10_-_27443155 | 27.97 |
ENST00000427324.1
ENST00000326799.3 |
YME1L1
|
YME1-like 1 ATPase |
| chr11_+_71938925 | 27.95 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr22_+_19467261 | 27.81 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr4_-_1713977 | 27.55 |
ENST00000318386.4
|
SLBP
|
stem-loop binding protein |
| chr4_-_1714037 | 26.66 |
ENST00000488267.1
ENST00000429429.2 ENST00000480936.1 |
SLBP
|
stem-loop binding protein |
| chr12_-_54071181 | 26.53 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr22_+_40742497 | 26.31 |
ENST00000216194.7
|
ADSL
|
adenylosuccinate lyase |
| chr1_-_43637915 | 26.22 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chrX_-_153707246 | 25.17 |
ENST00000407062.1
|
LAGE3
|
L antigen family, member 3 |
| chr1_+_156024552 | 24.96 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr22_+_40742512 | 24.49 |
ENST00000454266.2
ENST00000342312.6 |
ADSL
|
adenylosuccinate lyase |
| chr11_+_17298297 | 24.21 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chrX_+_131157322 | 24.13 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr2_-_215674374 | 24.08 |
ENST00000449967.2
ENST00000421162.1 ENST00000260947.4 |
BARD1
|
BRCA1 associated RING domain 1 |
| chrX_-_129299847 | 23.97 |
ENST00000319908.3
ENST00000287295.3 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr16_-_66864806 | 23.69 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chrX_+_131157290 | 23.22 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr1_+_156024525 | 23.20 |
ENST00000368305.4
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr14_+_64854958 | 23.11 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr13_-_113862401 | 23.00 |
ENST00000375459.1
|
PCID2
|
PCI domain containing 2 |
| chr11_+_17298255 | 22.66 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr1_-_6295975 | 22.35 |
ENST00000343813.5
ENST00000362035.3 |
ICMT
|
isoprenylcysteine carboxyl methyltransferase |
| chr10_-_17659357 | 22.18 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr7_-_140714739 | 21.30 |
ENST00000467334.1
ENST00000324787.5 |
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr10_-_43892279 | 21.19 |
ENST00000443950.2
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr16_-_103572 | 20.92 |
ENST00000293860.5
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr6_+_10748019 | 20.57 |
ENST00000543878.1
ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L
TMEM14B
|
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr4_-_84035868 | 20.54 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr3_-_25824925 | 20.54 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr10_-_27443294 | 20.52 |
ENST00000396296.3
ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1
|
YME1-like 1 ATPase |
| chr4_+_113558272 | 20.46 |
ENST00000509061.1
ENST00000508577.1 ENST00000513553.1 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr22_-_42086477 | 20.26 |
ENST00000402458.1
|
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr2_-_58468437 | 20.25 |
ENST00000403676.1
ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL
|
Fanconi anemia, complementation group L |
| chr15_+_66797627 | 20.03 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr4_-_84035905 | 19.90 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr18_-_47018869 | 19.66 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr7_+_75932863 | 19.44 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr5_+_34757309 | 19.23 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr18_-_47018897 | 19.20 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chrX_+_24072833 | 18.92 |
ENST00000253039.4
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr11_+_125496124 | 18.68 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chrX_-_153707545 | 18.49 |
ENST00000357360.4
|
LAGE3
|
L antigen family, member 3 |
| chr11_+_35201826 | 18.40 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr15_-_34635314 | 18.12 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chr6_+_10747986 | 18.04 |
ENST00000379542.5
|
TMEM14B
|
transmembrane protein 14B |
| chr22_+_20105259 | 18.00 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr19_+_57874835 | 17.57 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr5_+_133707252 | 17.54 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr4_+_57845043 | 16.94 |
ENST00000433463.1
ENST00000314595.5 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr5_+_31532373 | 16.90 |
ENST00000325366.9
ENST00000355907.3 ENST00000507818.2 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr16_+_103816 | 16.89 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr22_+_20105012 | 16.87 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr4_-_140223614 | 16.79 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr17_+_56769924 | 16.59 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr4_+_57845024 | 16.58 |
ENST00000431623.2
ENST00000441246.2 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr17_-_56084578 | 16.45 |
ENST00000582730.2
ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1
|
serine/arginine-rich splicing factor 1 |
| chr8_-_131455835 | 16.44 |
ENST00000518721.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr4_-_39367949 | 16.41 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr20_-_2451395 | 16.37 |
ENST00000339610.6
ENST00000381342.2 ENST00000438552.2 |
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr11_+_125495862 | 15.98 |
ENST00000428830.2
ENST00000544373.1 ENST00000527013.1 ENST00000526937.1 ENST00000534685.1 |
CHEK1
|
checkpoint kinase 1 |
| chr3_+_133293278 | 15.90 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr3_+_171758344 | 15.79 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr12_-_15942309 | 15.69 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr3_-_185542761 | 15.47 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr1_-_205719295 | 15.39 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr12_-_102455846 | 15.35 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr5_-_79950371 | 14.88 |
ENST00000511032.1
ENST00000504396.1 ENST00000505337.1 |
DHFR
|
dihydrofolate reductase |
| chr12_-_15942503 | 14.83 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr4_-_106629796 | 14.63 |
ENST00000416543.1
ENST00000515819.1 ENST00000420368.2 ENST00000503746.1 ENST00000340139.5 ENST00000433009.1 |
INTS12
|
integrator complex subunit 12 |
| chr14_-_100841670 | 14.57 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr18_-_47017956 | 14.55 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr3_-_185542817 | 14.44 |
ENST00000382199.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr1_-_26232522 | 14.30 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr3_-_125313934 | 14.16 |
ENST00000296220.5
|
OSBPL11
|
oxysterol binding protein-like 11 |
| chrX_+_41192595 | 13.95 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr4_-_140223670 | 13.93 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr1_-_24306835 | 13.85 |
ENST00000484146.2
|
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr7_+_128379346 | 13.63 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr1_+_158979792 | 13.54 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr3_+_197677047 | 13.28 |
ENST00000448864.1
|
RPL35A
|
ribosomal protein L35a |
| chr8_+_29953163 | 13.01 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr17_+_66243715 | 12.98 |
ENST00000359904.3
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr3_+_44803209 | 12.90 |
ENST00000326047.4
|
KIF15
|
kinesin family member 15 |
| chr7_+_128379449 | 12.89 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr5_+_82767583 | 12.89 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr8_+_48873453 | 12.09 |
ENST00000523944.1
|
MCM4
|
minichromosome maintenance complex component 4 |
| chr22_+_38071615 | 12.07 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr6_+_135502501 | 12.00 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr12_-_102455902 | 11.88 |
ENST00000240079.6
|
CCDC53
|
coiled-coil domain containing 53 |
| chr15_-_64673665 | 11.76 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr8_+_48873479 | 11.72 |
ENST00000262105.2
|
MCM4
|
minichromosome maintenance complex component 4 |
| chr17_+_61904766 | 11.64 |
ENST00000581842.1
ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5
|
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr2_+_238395803 | 11.55 |
ENST00000264605.3
|
MLPH
|
melanophilin |
| chr3_-_79816965 | 11.41 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr17_+_39975455 | 11.25 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr3_-_119396193 | 11.19 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr5_+_82767487 | 11.07 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chrX_+_52240504 | 11.06 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr17_-_9479128 | 10.97 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr6_-_111804905 | 10.68 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr14_-_20801427 | 10.59 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr2_-_130939115 | 10.46 |
ENST00000441135.1
ENST00000339679.7 ENST00000426662.2 ENST00000443958.2 ENST00000351288.6 ENST00000453750.1 ENST00000452225.2 |
SMPD4
|
sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) |
| chr2_-_86790593 | 10.42 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr2_+_63816295 | 10.38 |
ENST00000539945.1
ENST00000544381.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr12_+_56521840 | 10.37 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr17_-_29151794 | 10.30 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr12_-_123011536 | 10.11 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr12_+_3069037 | 9.84 |
ENST00000397122.2
|
TEAD4
|
TEA domain family member 4 |
| chr20_-_2644832 | 9.79 |
ENST00000380851.5
ENST00000380843.4 |
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr11_+_47236489 | 9.67 |
ENST00000256996.4
ENST00000378603.3 ENST00000378600.3 ENST00000378601.3 |
DDB2
|
damage-specific DNA binding protein 2, 48kDa |
| chr19_+_2270283 | 9.55 |
ENST00000588673.2
|
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr1_-_28241226 | 9.38 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr17_+_57642886 | 9.29 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr16_+_2732476 | 9.28 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr18_-_47018769 | 9.25 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr17_+_39975544 | 9.24 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr1_+_89150245 | 9.06 |
ENST00000370513.5
|
PKN2
|
protein kinase N2 |
| chrX_+_55026763 | 8.95 |
ENST00000374987.3
|
APEX2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr14_+_24899141 | 8.84 |
ENST00000556842.1
ENST00000553935.1 |
KHNYN
|
KH and NYN domain containing |
| chrX_-_10588459 | 8.79 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chrX_-_135338503 | 8.78 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr17_-_30668887 | 8.77 |
ENST00000581747.1
ENST00000583334.1 ENST00000580558.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr13_-_41345277 | 8.56 |
ENST00000323563.6
|
MRPS31
|
mitochondrial ribosomal protein S31 |
| chr3_+_124449213 | 8.40 |
ENST00000232607.2
ENST00000536109.1 ENST00000538242.1 ENST00000413078.2 |
UMPS
|
uridine monophosphate synthetase |
| chr14_-_51135036 | 8.09 |
ENST00000324679.4
|
SAV1
|
salvador homolog 1 (Drosophila) |
| chr2_+_238395879 | 8.08 |
ENST00000445024.2
ENST00000338530.4 ENST00000409373.1 |
MLPH
|
melanophilin |
| chr22_+_35653445 | 8.01 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr12_+_108079509 | 8.01 |
ENST00000412830.3
ENST00000547995.1 |
PWP1
|
PWP1 homolog (S. cerevisiae) |
| chr20_-_524340 | 7.61 |
ENST00000400227.3
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr12_+_1738363 | 7.53 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr1_-_6453399 | 7.43 |
ENST00000608083.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr7_-_56119238 | 7.42 |
ENST00000275605.3
ENST00000395471.3 |
PSPH
|
phosphoserine phosphatase |
| chr12_+_123011776 | 7.22 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr17_+_76165213 | 7.14 |
ENST00000590201.1
|
SYNGR2
|
synaptogyrin 2 |
| chr1_+_201798269 | 7.05 |
ENST00000361565.4
|
IPO9
|
importin 9 |
| chr12_+_111843749 | 7.03 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr13_-_30169807 | 6.97 |
ENST00000380752.5
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr7_+_856246 | 6.97 |
ENST00000389574.3
ENST00000457378.2 ENST00000452783.2 ENST00000435699.1 ENST00000440380.1 ENST00000439679.1 ENST00000424128.1 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chrX_-_10588595 | 6.65 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr2_+_232575128 | 6.59 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr3_-_194393206 | 6.58 |
ENST00000265245.5
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr15_+_44119159 | 6.46 |
ENST00000263795.6
ENST00000381246.2 ENST00000452115.1 |
WDR76
|
WD repeat domain 76 |
| chr6_+_43603552 | 6.45 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr8_+_29952914 | 6.36 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr1_-_89458287 | 6.34 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr6_-_2962331 | 6.21 |
ENST00000380524.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chrX_-_53449593 | 6.08 |
ENST00000375340.6
ENST00000322213.4 |
SMC1A
|
structural maintenance of chromosomes 1A |
| chr2_+_130939235 | 6.07 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr17_-_46623441 | 5.90 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr6_-_32157947 | 5.64 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr8_-_81083731 | 5.52 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr19_-_19249255 | 5.31 |
ENST00000587583.2
ENST00000450333.2 ENST00000587096.1 ENST00000162044.9 ENST00000592369.1 ENST00000587915.1 |
TMEM161A
|
transmembrane protein 161A |
| chr12_-_123011476 | 5.17 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr21_-_43346790 | 5.17 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr19_-_47291843 | 5.14 |
ENST00000542575.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr1_+_33283043 | 5.11 |
ENST00000373476.1
ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP
|
S100P binding protein |
| chr11_+_134201911 | 5.07 |
ENST00000389881.3
|
GLB1L2
|
galactosidase, beta 1-like 2 |
| chr20_+_60174827 | 5.04 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr6_+_26251835 | 4.91 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr7_-_22259845 | 4.91 |
ENST00000420196.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr1_+_150245099 | 4.81 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr4_-_4543700 | 4.76 |
ENST00000505286.1
ENST00000306200.2 |
STX18
|
syntaxin 18 |
| chr1_-_21503337 | 4.73 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr19_+_50887585 | 4.49 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr2_+_63816087 | 4.46 |
ENST00000409908.1
ENST00000442225.1 ENST00000409476.1 ENST00000436321.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr4_-_129209944 | 4.41 |
ENST00000520121.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr17_+_37894570 | 4.38 |
ENST00000394211.3
|
GRB7
|
growth factor receptor-bound protein 7 |
| chr15_+_40731920 | 4.27 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr2_-_70780572 | 4.11 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr15_-_89456630 | 4.10 |
ENST00000268150.8
|
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chrX_-_54522558 | 4.06 |
ENST00000375135.3
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr11_+_65029233 | 3.96 |
ENST00000265465.3
|
POLA2
|
polymerase (DNA directed), alpha 2, accessory subunit |
| chr10_-_121632266 | 3.94 |
ENST00000360003.3
ENST00000369077.3 |
MCMBP
|
minichromosome maintenance complex binding protein |
| chr14_+_105992906 | 3.83 |
ENST00000392519.2
|
TMEM121
|
transmembrane protein 121 |
| chr2_+_132233664 | 3.80 |
ENST00000321253.6
|
TUBA3D
|
tubulin, alpha 3d |
| chr16_-_71264558 | 3.74 |
ENST00000448089.2
ENST00000393550.2 ENST00000448691.1 ENST00000393567.2 ENST00000321489.5 ENST00000539973.1 ENST00000288168.10 ENST00000545267.1 ENST00000541601.1 ENST00000538248.1 |
HYDIN
|
HYDIN, axonemal central pair apparatus protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 30.4 | 91.2 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 16.9 | 50.8 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 11.7 | 116.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 11.3 | 67.9 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 11.1 | 66.6 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 10.8 | 54.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 10.2 | 30.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 9.3 | 28.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 9.3 | 27.8 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 8.1 | 48.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 7.6 | 30.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 6.8 | 20.5 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 6.0 | 60.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 5.8 | 17.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 5.8 | 34.7 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 4.9 | 19.4 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 4.7 | 38.0 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 4.7 | 23.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 4.6 | 23.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 3.8 | 15.4 | GO:0019046 | release from viral latency(GO:0019046) |
| 3.7 | 33.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 3.6 | 18.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 3.4 | 24.0 | GO:1904044 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) response to aldosterone(GO:1904044) |
| 3.3 | 16.6 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 3.3 | 16.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 3.2 | 19.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 3.0 | 12.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 3.0 | 24.1 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 2.9 | 14.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 2.9 | 66.7 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 2.9 | 14.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 2.8 | 2.8 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 2.8 | 8.4 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 2.8 | 14.0 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 2.7 | 8.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 2.3 | 18.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.2 | 40.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 2.0 | 28.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 2.0 | 14.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 2.0 | 5.9 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 1.9 | 9.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.9 | 70.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 1.9 | 7.4 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 1.9 | 31.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 1.8 | 5.3 | GO:0050923 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.7 | 7.0 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 1.7 | 7.0 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 1.5 | 6.1 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 1.4 | 31.3 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 1.4 | 9.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 1.4 | 9.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.4 | 1.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.4 | 4.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.3 | 19.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 1.3 | 10.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 1.3 | 48.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 1.3 | 11.6 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 1.3 | 5.1 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 1.3 | 31.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 1.2 | 18.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 1.2 | 14.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 1.2 | 10.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.2 | 15.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 1.2 | 2.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 1.1 | 20.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 1.0 | 10.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.0 | 3.0 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 1.0 | 24.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.9 | 28.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.9 | 71.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.9 | 11.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.8 | 26.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.8 | 4.8 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.8 | 10.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.8 | 16.4 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.7 | 20.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.7 | 35.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.7 | 9.7 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.7 | 4.4 | GO:0071407 | cellular response to organic cyclic compound(GO:0071407) |
| 0.7 | 14.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.6 | 10.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.6 | 20.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.6 | 13.7 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.6 | 1.7 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.5 | 0.5 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.5 | 2.1 | GO:1904457 | glucosylceramide catabolic process(GO:0006680) termination of signal transduction(GO:0023021) beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 1.9 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.5 | 59.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.4 | 1.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 30.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 32.2 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.4 | 7.0 | GO:0051170 | nuclear import(GO:0051170) |
| 0.4 | 5.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.4 | 29.9 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.4 | 43.7 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.4 | 8.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.4 | 6.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.4 | 14.5 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.3 | 27.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.3 | 4.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.3 | 13.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.3 | 4.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.3 | 0.9 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.3 | 6.6 | GO:0006351 | transcription, DNA-templated(GO:0006351) nucleic acid-templated transcription(GO:0097659) |
| 0.3 | 2.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.3 | 4.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.3 | 22.4 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.3 | 1.9 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.3 | 21.2 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.3 | 4.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.3 | 29.1 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.2 | 1.9 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 3.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 9.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.2 | 5.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 9.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.2 | 5.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 23.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.2 | 5.3 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.2 | 1.8 | GO:0055091 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) phospholipid homeostasis(GO:0055091) |
| 0.1 | 12.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 12.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 10.3 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 4.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 3.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 9.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 3.7 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 12.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 2.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 20.3 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 3.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 5.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 6.6 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.1 | 8.9 | GO:0044764 | multi-organism cellular process(GO:0044764) |
| 0.1 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 4.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 7.7 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 1.7 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.7 | 66.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 14.5 | 188.0 | GO:0042555 | MCM complex(GO:0042555) |
| 10.7 | 32.2 | GO:0031523 | Myb complex(GO:0031523) |
| 9.6 | 48.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 9.3 | 27.8 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 8.8 | 70.6 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 8.7 | 43.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 7.9 | 31.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 6.9 | 34.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 6.8 | 27.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 6.3 | 18.9 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 6.0 | 24.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 4.7 | 28.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 4.3 | 12.9 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 4.0 | 67.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 3.5 | 17.5 | GO:0033503 | HULC complex(GO:0033503) |
| 3.0 | 18.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 2.9 | 20.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 2.6 | 18.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 2.4 | 26.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 2.3 | 16.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 2.0 | 30.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.9 | 58.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 1.7 | 23.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 1.6 | 14.6 | GO:0032039 | integrator complex(GO:0032039) |
| 1.5 | 26.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.2 | 11.1 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 1.2 | 9.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.2 | 10.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 1.1 | 8.0 | GO:0016589 | NURF complex(GO:0016589) |
| 1.1 | 4.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.1 | 33.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 1.1 | 29.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 1.0 | 6.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 1.0 | 6.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 1.0 | 11.6 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.8 | 23.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.8 | 10.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.7 | 10.4 | GO:0043601 | replisome(GO:0030894) nuclear replisome(GO:0043601) |
| 0.7 | 16.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.7 | 1.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.7 | 72.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 10.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.6 | 1.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.6 | 16.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.6 | 16.4 | GO:0002102 | podosome(GO:0002102) |
| 0.6 | 14.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.5 | 7.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 71.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.5 | 4.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.5 | 30.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 30.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.4 | 61.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.4 | 55.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.4 | 40.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.4 | 0.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 15.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 1.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.3 | 2.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 46.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.3 | 15.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 1.8 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 22.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 6.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 4.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 25.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.3 | 15.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 31.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 15.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 2.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 4.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 3.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.2 | 6.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 63.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 12.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 2.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 4.9 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.1 | 7.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 22.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 4.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 9.1 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 13.3 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 1.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 3.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 5.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 31.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 14.7 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 10.4 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 2.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 2.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 14.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.7 | 66.7 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 15.0 | 60.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 14.1 | 70.6 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 11.2 | 123.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 11.0 | 33.1 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 7.8 | 132.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 7.7 | 23.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 6.5 | 19.4 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 6.3 | 50.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 6.1 | 48.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 5.8 | 34.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 5.7 | 28.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 5.0 | 35.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 4.5 | 22.4 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 4.0 | 12.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 3.0 | 18.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 3.0 | 9.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 3.0 | 14.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 2.9 | 14.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 2.8 | 28.0 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 2.5 | 17.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 2.5 | 14.9 | GO:0051870 | methotrexate binding(GO:0051870) |
| 2.4 | 9.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 2.1 | 94.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 2.1 | 16.6 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 2.0 | 10.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 1.9 | 9.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.9 | 11.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.9 | 11.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 1.8 | 23.7 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 1.7 | 7.0 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 1.7 | 87.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 1.5 | 6.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 1.4 | 28.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 1.4 | 20.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 1.2 | 36.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.1 | 65.6 | GO:0070888 | E-box binding(GO:0070888) |
| 1.1 | 31.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 1.1 | 11.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.0 | 10.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.0 | 11.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.0 | 26.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.9 | 10.4 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.9 | 29.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.8 | 39.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.7 | 7.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.7 | 5.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.7 | 21.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.7 | 20.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.7 | 19.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.7 | 10.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.6 | 5.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.6 | 15.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.6 | 1.8 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.6 | 30.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 2.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.5 | 56.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.5 | 2.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.5 | 4.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.5 | 102.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.5 | 9.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 1.9 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.5 | 30.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.4 | 22.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.4 | 7.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 17.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.4 | 9.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.4 | 4.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 23.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 68.1 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.2 | 7.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 13.3 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.2 | 9.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 3.6 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 13.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 2.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 8.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 6.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 9.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.7 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.9 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 2.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 25.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 4.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 19.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 58.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 8.2 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 4.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 5.5 | GO:0008289 | lipid binding(GO:0008289) |
| 0.1 | 5.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 13.0 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.1 | 24.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 8.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 4.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 8.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 5.3 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 157.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 1.4 | 20.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.4 | 77.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 1.2 | 28.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 1.1 | 128.9 | PID E2F PATHWAY | E2F transcription factor network |
| 1.1 | 44.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 1.0 | 46.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.8 | 19.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.8 | 14.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.7 | 16.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.6 | 40.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.6 | 18.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 28.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.5 | 30.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.4 | 22.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 31.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.4 | 13.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 15.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.3 | 16.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 23.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 7.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 9.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 4.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 4.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 5.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 9.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 6.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 9.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.2 | 211.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 4.4 | 70.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 3.3 | 39.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 3.0 | 81.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 3.0 | 60.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 3.0 | 50.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.9 | 23.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 1.9 | 32.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 1.7 | 10.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.6 | 73.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 1.5 | 66.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 1.4 | 33.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 1.1 | 24.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.0 | 33.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.0 | 18.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.0 | 9.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.9 | 31.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.8 | 18.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.7 | 112.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.6 | 28.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.5 | 10.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.5 | 11.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.5 | 91.6 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.5 | 6.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 9.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.4 | 14.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 31.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.4 | 11.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 10.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 30.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 23.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 8.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 12.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 9.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 15.8 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.3 | 27.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.3 | 4.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 5.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 5.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 19.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 3.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 6.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 15.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 10.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.2 | 26.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 5.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 5.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 19.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 3.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 5.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 10.6 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.1 | 1.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |